3E79
 
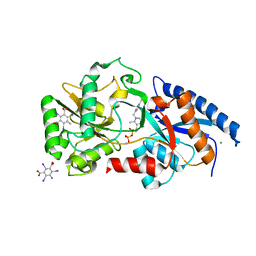 | | Structure determination of the cancer-associated Mycoplasma hyorhinis protein Mh-p37 | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sippel, K.H, Robbins, A.H, Reutzel, R, Domsic, J, McKenna, R. | | Deposit date: | 2008-08-18 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure determination of the cancer-associated Mycoplasma hyorhinis protein Mh-p37.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3EM3
 
 | |
3FTU
 
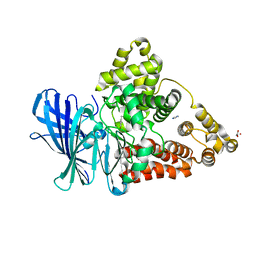 | | Leukotriene A4 hydrolase in complex with dihydroresveratrol | | Descriptor: | 5-[2-(4-hydroxyphenyl)ethyl]benzene-1,3-diol, ACETATE ION, IMIDAZOLE, ... | | Authors: | Davies, D.R. | | Deposit date: | 2009-01-13 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of leukotriene A4 hydrolase inhibitors using metabolomics biased fragment crystallography.
J.Med.Chem., 52, 2009
|
|
3FSI
 
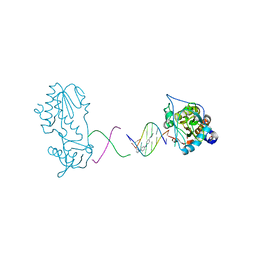 | | Crystal structure of a trypanocidal 4,4'-Bis(imidazolinylamino)diphenylamine bound to DNA | | Descriptor: | 5'-D(*CP*TP*TP*AP*AP*TP*TP*C)-3', 5'-D(P*GP*AP*AP*TP*TP*AP*AP*G)-3', ACETATE ION, ... | | Authors: | Glass, L.S, Georgiadis, M.M, Goodwin, K.D. | | Deposit date: | 2009-01-09 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a trypanocidal 4,4'-bis(imidazolinylamino)diphenylamine bound to DNA.
Biochemistry, 48, 2009
|
|
3EPF
 
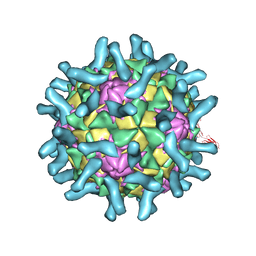 | | CryoEM structure of poliovirus receptor bound to poliovirus type 2 | | Descriptor: | 1[2-CHLORO-4-METHOXY-PHENYL-OXYMETHYL]-4-[2,6-DICHLORO-PHENYL-OXYMETHYL]-BENZENE, MYRISTIC ACID, Poliovirus receptor, ... | | Authors: | Zhang, P, Mueller, S, Morais, M.C, Bator, C.M, Bowman, V.D, Hafenstein, S, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-11 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3E9Y
 
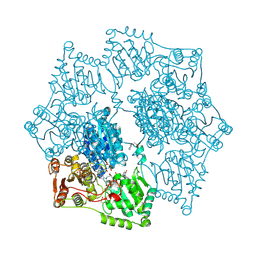 | | Arabidopsis thaliana acetohydroxyacid synthase in complex with monosulfuron | | Descriptor: | 2-[(2E)-3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-HYDROXYETHYLIDENE)-4-METHYL-2,3-DIHYDRO-1,3-THIAZOL-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Acetolactate synthase, ... | | Authors: | Guddat, L.W, Duggleby, R.G, Wang, J.-G, Li, Z.-M. | | Deposit date: | 2008-08-24 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of two novel sulfonylurea herbicides in complex with Arabidopsis thaliana acetohydroxyacid synthase.
Febs J., 276, 2009
|
|
3H3C
 
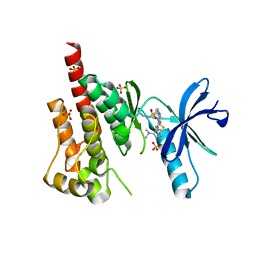 | | Crystal structure of PYK2 in complex with Sulfoximine-substituted trifluoromethylpyrimidine analog | | Descriptor: | 4-{[4-{[(1R,2R)-2-(dimethylamino)cyclopentyl]amino}-5-(trifluoromethyl)pyrimidin-2-yl]amino}-N-methylbenzenesulfonamide, Protein tyrosine kinase 2 beta, SULFATE ION | | Authors: | Han, S, Mistry, A. | | Deposit date: | 2009-04-16 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sulfoximine-substituted trifluoromethylpyrimidine analogs as inhibitors of proline-rich tyrosine kinase 2 (PYK2) show reduced hERG activity.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3CKT
 
 | | HIV-1 protease in complex with a dimethylallyl decorated pyrrolidine based inhibitor (orthorombic space group) | | Descriptor: | (3S,4S),-3,4-Bis-[(4-carbamoyl-benzensulfonyl)-(3-methyl-but-2-enyl)-amino]-pyrrolidine, CHLORIDE ION, Protease | | Authors: | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2008-03-17 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Two Solutions for the Same Problem: Multiple Binding Modes of Pyrrolidine-Based HIV-1 Protease Inhibitors
J.Mol.Biol., 410, 2011
|
|
3CIC
 
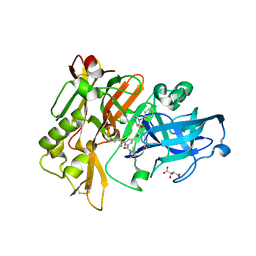 | | Structure of BACE Bound to SCH709583 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2S)-2-[(2S)-4-benzyl-3-oxopiperazin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2008-03-11 | | Release date: | 2008-06-10 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3GT4
 
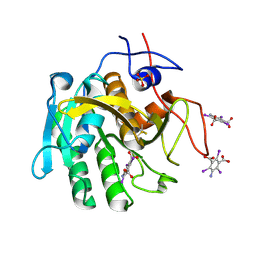 | | Structure of proteinase K with the magic triangle I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, SULFATE ION, proteinase K | | Authors: | Beck, T, Gruene, T, Sheldrick, G.M. | | Deposit date: | 2009-03-27 | | Release date: | 2009-04-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The magic triangle goes MAD: experimental phasing with a bromine derivative
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3GT3
 
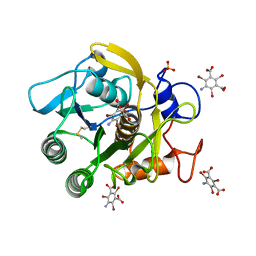 | | Structure of proteinase K with the mad triangle B3C | | Descriptor: | 5-amino-2,4,6-tribromobenzene-1,3-dicarboxylic acid, Proteinase K, SULFATE ION | | Authors: | Beck, T, Gruene, T, Sheldrick, G.M. | | Deposit date: | 2009-03-27 | | Release date: | 2009-04-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The magic triangle goes MAD: experimental phasing with a bromine derivative
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5XLZ
 
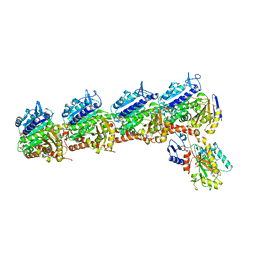 | |
5Y4S
 
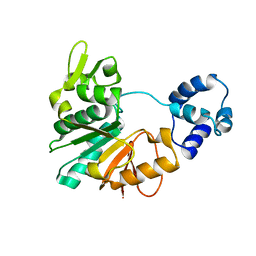 | | Structure of a methyltransferase complex | | Descriptor: | Chemotaxis protein methyltransferase 1 | | Authors: | Yan, X, Xin, L, Tan, Y.J, Jin, S, Liang, Z.X, Gao, Y.G. | | Deposit date: | 2017-08-04 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.405 Å) | | Cite: | Structural analyses unravel the molecular mechanism of cyclic di-GMP regulation of bacterial chemotaxis via a PilZ adaptor protein.
J. Biol. Chem., 293, 2018
|
|
5YE2
 
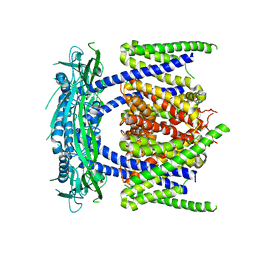 | |
5YEV
 
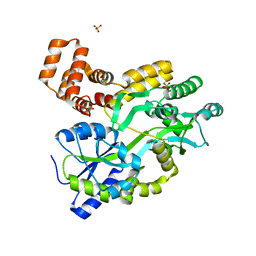 | | Murine DR3 death domain | | Descriptor: | SULFATE ION, TNFRSF25 death domain | | Authors: | Yin, X, Jin, T. | | Deposit date: | 2017-09-19 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
5YE1
 
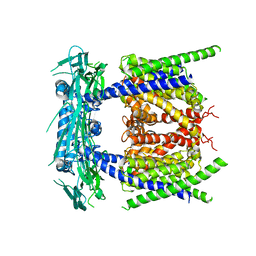 | |
5YGP
 
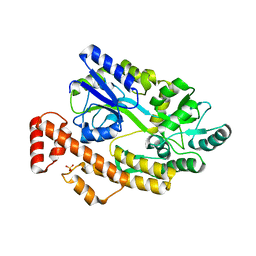 | | Human TNFRSF25 death domain mutant-D412E | | Descriptor: | SULFATE ION, TNFRSF25 death domain, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yin, X, Jin, T.C. | | Deposit date: | 2017-09-25 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
5XXY
 
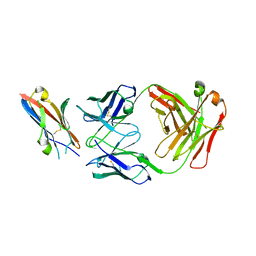 | |
5Y4R
 
 | | Structure of a methyltransferase complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Chemotaxis protein methyltransferase 1, Cyclic diguanosine monophosphate-binding protein PA4608, ... | | Authors: | Yan, X, Xin, L, Tan, Y.J, Jin, S, Liang, Z.X, Gao, Y.G. | | Deposit date: | 2017-08-04 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Structural analyses unravel the molecular mechanism of cyclic di-GMP regulation of bacterial chemotaxis via a PilZ adaptor protein.
J. Biol. Chem., 293, 2018
|
|
5WUD
 
 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
5YI8
 
 | |
5YD0
 
 | | Crystal structure of Schlafen 13 (SLFN13) N'-domain | | Descriptor: | Schlafen 8, ZINC ION | | Authors: | Yang, J.-Y, Gao, S. | | Deposit date: | 2017-09-09 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.182 Å) | | Cite: | Structure of Schlafen13 reveals a new class of tRNA/rRNA- targeting RNase engaged in translational control
Nat Commun, 9, 2018
|
|
5YKF
 
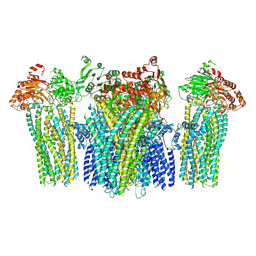 | |
5WT1
 
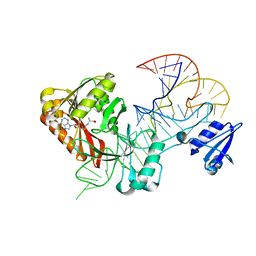 | |
5WPQ
 
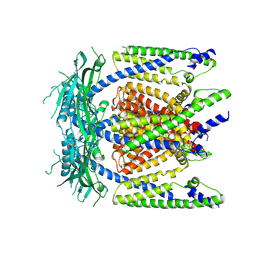 | | Cryo-EM structure of mammalian endolysosomal TRPML1 channel in nanodiscs in closed I conformation at 3.64 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-1, SODIUM ION | | Authors: | Chen, Q, She, J, Guo, J, Bai, X, Jiang, Y. | | Deposit date: | 2017-08-07 | | Release date: | 2017-10-18 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structure of mammalian endolysosomal TRPML1 channel in nanodiscs.
Nature, 550, 2017
|
|
