5WZW
 
 | | Crystal structure of human secreted phospholipase A2 group IIE with LY311727 | | Descriptor: | (3-{[3-(2-amino-2-oxoethyl)-1-benzyl-2-ethyl-1H-indol-5-yl]oxy}propyl)phosphonic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
8I3V
 
 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of the antidepressant escitalopram in an inward-open state at resolution of 2.85 angstrom. | | Descriptor: | (1S)-1-[3-(dimethylamino)propyl]-1-(4-fluorophenyl)-1,3-dihydro-2-benzofuran-5-carbonitrile, CHLORIDE ION, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2023-01-18 | | Release date: | 2024-07-03 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 632, 2024
|
|
7U0S
 
 | | Crystal Structure of FK506-binding protein 1A from Aspergillus fumigatus Bound to Ascomycin | | Descriptor: | (3S,4R,5S,8R,9E,12S,14S,15R,16S,18R,19R,22R,26aS)-8-ethyl-5,19-dihydroxy-3-{(1E)-1-[(1R,3R,4R)-4-hydroxy-3-methoxycyclohexyl]prop-1-en-2-yl}-14,16-dimethoxy-4,10,12,18-tetramethyl-5,6,8,11,12,13,14,15,16,17,18,19,24,25,26,26a-hexadecahydro-3H-15,19-epoxypyrido[2,1-c][1,4]oxazacyclotricosine-1,7,20,21(4H,23H)-tetrone, ACETATE ION, FK506-binding protein 1A, ... | | Authors: | DeBouver, N.D, Fox III, D, Hoy, M.J, Heitman, J, Lorimer, D.D, Horanyi, P.S, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-02-18 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Synthesis of FK506 and FK520 Analogs with Increased Selectivity Exhibit In Vivo Therapeutic Efficacy against Cryptococcus.
Mbio, 13, 2022
|
|
8CSP
 
 | | Human mitochondrial small subunit assembly intermediate (State A) | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Harper, N.J, Burnside, C, Klinge, S. | | Deposit date: | 2022-05-13 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
6DMI
 
 | | A multiconformer ligand model of 5T5 bound to BACE-1 | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, SODIUM ION, ... | | Authors: | Hudson, B.M, van Zundert, G, Keedy, D.A, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
8CSQ
 
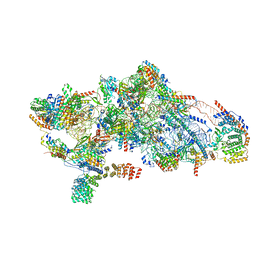 | | Human mitochondrial small subunit assembly intermediate (State B) | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Harper, N.J, Burnside, C, Klinge, S. | | Deposit date: | 2022-05-13 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
5AHT
 
 | |
9HJH
 
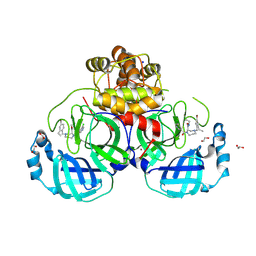 | | Structure of compound 1 bound to SARS-CoV-2 main protease | | Descriptor: | (2~{R})-4-[(4-bromanyl-2-ethyl-phenyl)methyl]-1-(5-chloranylpyridin-3-yl)carbonyl-~{N}-ethyl-1,4-diazepane-2-carboxamide, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Mac Sweeney, A, Hazemann, J. | | Deposit date: | 2024-11-29 | | Release date: | 2024-12-11 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Accelerating the Hit-To-Lead Optimization of a SARS-CoV-2 Mpro Inhibitor Series by Combining High-Throughput Medicinal Chemistry and Computational Simulations.
J.Med.Chem., 68, 2025
|
|
5WAF
 
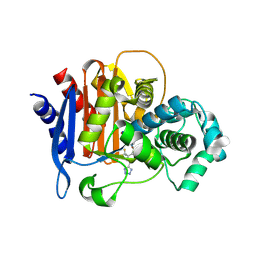 | | ADC-7 in complex with boronic acid transition state inhibitor CR192 | | Descriptor: | Beta-lactamase, phosphonooxy-[[[4-(1~{H}-1,2,3,4-tetrazol-5-yl)-2-(trifluoromethyl)phenyl]sulfonylamino]methyl]borinic acid | | Authors: | Powers, R.A, Wallar, B.J. | | Deposit date: | 2017-06-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure-Based Analysis of Boronic Acids as Inhibitors of Acinetobacter-Derived Cephalosporinase-7, a Unique Class C beta-Lactamase.
ACS Infect Dis, 4, 2018
|
|
1AVA
 
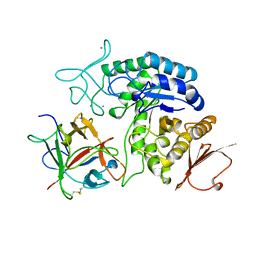 | | AMY2/BASI PROTEIN-PROTEIN COMPLEX FROM BARLEY SEED | | Descriptor: | BARLEY ALPHA-AMYLASE 2(CV MENUET), BARLEY ALPHA-AMYLASE/SUBTILISIN INHIBITOR, CALCIUM ION | | Authors: | Vallee, F, Kadziola, A, Bourne, Y, Juy, M, Svensson, B, Haser, R. | | Deposit date: | 1997-09-15 | | Release date: | 1999-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Barley alpha-amylase bound to its endogenous protein inhibitor BASI: crystal structure of the complex at 1.9 A resolution.
Structure, 6, 1998
|
|
5CAP
 
 | | EGFR kinase domain mutant "TMLR" with compound 30 | | Descriptor: | 2-methyl-N-[2-(2-methyl-2-methylsulfonyl-propoxy)pyrimidin-4-yl]-1-propan-2-yl-imidazo[4,5-c]pyridin-6-amine, Epidermal growth factor receptor, SULFATE ION | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2015-06-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Noncovalent Mutant Selective Epidermal Growth Factor Receptor Inhibitors: A Lead Optimization Case Study.
J.Med.Chem., 58, 2015
|
|
4U2Z
 
 | | X-ray crystal structure of an Sco GlgEI-V279S/1,2,2-trifluromaltose complex | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, alpha-D-glucopyranose-(1-4)-2-deoxy-2,2-difluoro-alpha-D-arabino-hexopyranosyl fluoride | | Authors: | Ronning, D.R, Lindenberger, J.J. | | Deposit date: | 2014-07-18 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Synthesis of 2-deoxy-2,2-difluoro-alpha-maltosyl fluoride and its X-ray structure in complex with Streptomyces coelicolor GlgEI-V279S.
Org.Biomol.Chem., 13, 2015
|
|
6D6W
 
 | | Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate | | Descriptor: | Beta-galactosidase/beta-glucuronidase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2018-04-23 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three structurally and functionally distinct beta-glucuronidases from the human gut microbeBacteroides uniformis.
J. Biol. Chem., 293, 2018
|
|
5JHB
 
 | | Structure of Phosphoinositide 3-kinase gamma (PI3K) bound to the potent inhibitor PIKin3 | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION, [1-{4-[6-amino-4-(trifluoromethyl)pyridin-3-yl]-6-(morpholin-4-yl)-1,3,5-triazin-2-yl}-3-(chloromethyl)azetidin-3-yl]methanol | | Authors: | Burke, J.E, Inglis, A.J, Williams, R.L. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Deconvolution of Buparlisib's mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention.
Nat Commun, 8, 2017
|
|
5A4J
 
 | | Crystal structure of FTHFS1 from T.acetoxydans Re1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, D(-)-TARTARIC ACID, ... | | Authors: | Bergdahl, R, Jacobson, F, Muller, B, Mikkelsen, N, Schurer, A, Sandgren, M. | | Deposit date: | 2015-06-10 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Characterization, Crystallization and Three- Dimensional Structures of Formyltetrahydrofolate Synthetase (Fthfs) from the Syntrophic Acetate Oxidising Bacterium Tepidanaerobacter Acetatoxydans Re1
To be Published
|
|
7U5G
 
 | | ACS122 Fab | | Descriptor: | ACS122 Fab Heavy chain, ACS122 Fab Light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-03-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complementary antibody lineages achieve neutralization breadth in an HIV-1 infected elite neutralizer.
Plos Pathog., 18, 2022
|
|
7LMO
 
 | | Structure of full-length human lambda-6A light chain JTO in complex with stabilizer 34 [3-(2-(7-(diethylamino)-4-methyl-2-oxo-2H-chromen-3-yl)ethyl)-7-(1H-imidazole-5-carbonyl)-1,3,7-triazaspiro[4.4]nonane-2,4-dione] | | Descriptor: | (5~{R})-3-[2-[7-(diethylamino)-4-methyl-2-oxidanylidene-chromen-3-yl]ethyl]-7-(1~{H}-imidazol-4-ylcarbonyl)-1,3,7-triazaspiro[4.4]nonane-2,4-dione, JTO light chain, PHOSPHATE ION | | Authors: | Yan, N.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2021-02-05 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of Potent Coumarin-Based Kinetic Stabilizers of Amyloidogenic Immunoglobulin Light Chains Using Structure-Based Design.
J.Med.Chem., 64, 2021
|
|
6FEQ
 
 | | Structure of inhibitor-bound ABCG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3(Fab) heavy chain variable domain, 5D3(Fab) light chain variable domain, ... | | Authors: | Jackson, S.M, Manolaridis, I, Kowal, J, Zechner, M, Altmann, K.H, Locher, K.P. | | Deposit date: | 2018-01-03 | | Release date: | 2018-04-11 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of small-molecule inhibition of human multidrug transporter ABCG2.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7LMP
 
 | | Structure of full-length human lambda-6A light chain JTO in complex with stabilizer 36 [3-(2-(7-(diethylamino)-4-methyl-2-oxo-2H-chromen-3-yl)ethyl)-8-(1H-imidazole-4-carbonyl)-1,3,8-triazaspiro[4.5]decane-2,4-dione] | | Descriptor: | 3-[2-[7-(diethylamino)-4-methyl-2-oxidanylidene-chromen-3-yl]ethyl]-8-(1~{H}-imidazol-5-ylcarbonyl)-1,3,8-triazaspiro[4.5]decane-2,4-dione, JTO light chain, PHOSPHATE ION | | Authors: | Yan, N.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2021-02-05 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of Potent Coumarin-Based Kinetic Stabilizers of Amyloidogenic Immunoglobulin Light Chains Using Structure-Based Design.
J.Med.Chem., 64, 2021
|
|
8JU9
 
 | |
7TS0
 
 | | Cryo-EM structure of corticotropin releasing factor receptor 2 bound to Urocortin 1 and coupled with heterotrimeric Go protein | | Descriptor: | Corticotropin-releasing factor receptor 2,Corticotropin-releasing factor receptor 2,Corticotropin-releasing factor receptor 2,Corticotropin-releasing factor receptor 2,Human corticotropin releasing factor receptor 2, Dominant negative Go alpha subunit, G protein gamma subunit, ... | | Authors: | Zhao, L.-H, Lin, J, Mao, C, Zhou, X.E, Ji, S, Shen, D, Xiao, P, Melcher, K, Zhang, Y, Yu, X, Xu, H.E. | | Deposit date: | 2022-01-31 | | Release date: | 2022-11-09 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure insights into selective coupling of G protein subtypes by a class B G protein-coupled receptor.
Nat Commun, 13, 2022
|
|
7TRY
 
 | | Cryo-EM structure of corticotropin releasing factor receptor 2 bound to Urocortin 1 and coupled with heterotrimeric G11 protein | | Descriptor: | Corticotropin-releasing factor receptor 2, G protein gamma subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, L.-H, Lin, J, Mao, C, Zhou, X.E, Ji, S, Shen, D, Xiao, P, Melcher, K, Zhang, Y, Yu, X, Xu, H.E. | | Deposit date: | 2022-01-31 | | Release date: | 2022-11-09 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure insights into selective coupling of G protein subtypes by a class B G protein-coupled receptor.
Nat Commun, 13, 2022
|
|
5WZX
 
 | | Structural basis for a pentacyclic oleanane-type triterpenoid as a ligand of FXR | | Descriptor: | (4aR,6aR,6aS,6bS,8aS,9R,12aR,14bR)-2,2,6a,6b,9,12a-hexamethyl-10-oxidanylidene-1,3,4,5,6,6a,7,8,8a,9,11,12,13,14b-tetradecahydropicene-4a-carboxylic acid, (R,R)-2,3-BUTANEDIOL, Bile acid receptor, ... | | Authors: | Lu, Y, Li, Y. | | Deposit date: | 2017-01-19 | | Release date: | 2018-01-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Identification of an Oleanane-Type Triterpene Hedragonic Acid as a Novel Farnesoid X Receptor Ligand with Liver Protective Effects and Anti-inflammatory Activity
Mol. Pharmacol., 93, 2018
|
|
5AD3
 
 | | Bivalent binding to BET bromodomains | | Descriptor: | 3-methoxy-N-[2-[4-[1-(3-methoxy-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidyl]phenoxy]ethyl]-N-methyl-[1,2,4]triazolo[4,3-b]pyridazin-6-amine, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Waring, M.J, Chen, H, Rabow, A.A, Walker, G, Bobby, R, Boiko, S, Bradbury, R.H, Callis, R, Dale, I, Daniels, D, Flavell, L, Holdgate, G, Jowitt, T.A, Kikhney, A, McAlister, M, Ogg, D, Patel, J, Petteruti, P, Robb, G.R, Robers, M, Stratton, N, Svergun, D.I, Wang, W, Whittaker, D. | | Deposit date: | 2015-08-19 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Potent and Selective Bivalent Inhibitors of Bet Bromodomains
Nat.Chem.Biol., 12, 2016
|
|
6V1K
 
 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to BI7273 | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-[(dimethylamino)methyl]-3,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, Bromodomain-containing protein 4 | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
