6PNP
 
 | | Crystal structure of the splice insert-free neurexin-1 LNS2 domain in complex with neurexophilin-1 | | Descriptor: | Neurexin-1, Neurexophilin-1 | | Authors: | Wilson, S.C, White, K.I, Zhou, Q, Brunger, A.T. | | Deposit date: | 2019-07-02 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of neurexophilin-neurexin complexes reveal a regulatory mechanism of alternative splicing.
Embo J., 38, 2019
|
|
6IVK
 
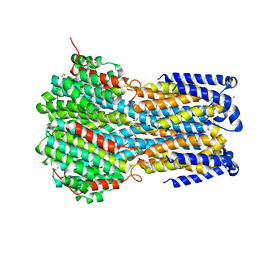 | | Crystal structure of a membrane protein G175A | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
2RP1
 
 | |
3KS5
 
 | |
6FMI
 
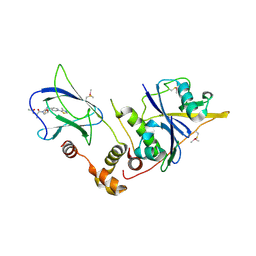 | | pVHL:EloB:EloC in complex with N-((S)-1-((2S,4R)-4-Hydroxy-2-((4-(4-methylthiazol-5-yl)benzyl)carbamothioyl) pyrrolidin-1-yl)-1-oxopropan-2-yl)acetamide (ligand 2) | | Descriptor: | Elongin-B, Elongin-C, von Hippel-Lindau disease tumor suppressor, ... | | Authors: | Soares, P, Lucas, X, Ciulli, A. | | Deposit date: | 2018-01-31 | | Release date: | 2018-04-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Thioamide substitution to probe the hydroxyproline recognition of VHL ligands.
Bioorg. Med. Chem., 26, 2018
|
|
8YZ6
 
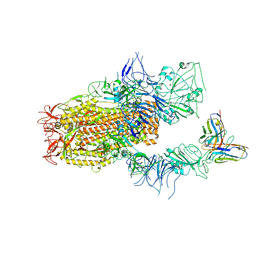 | | SARS-CoV-2 Spike (BA.1) in complex with Fab of JH-8B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain of JH-8B, ... | | Authors: | Chen, X, Wu, Y.-M. | | Deposit date: | 2024-04-06 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
3ZRH
 
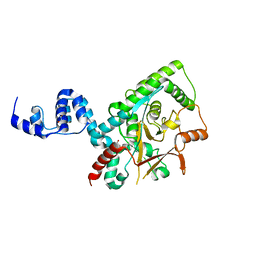 | | Crystal structure of the Lys29, Lys33-linkage-specific TRABID OTU deubiquitinase domain reveals an Ankyrin-repeat ubiquitin binding domain (AnkUBD) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, UBIQUITIN THIOESTERASE ZRANB1 | | Authors: | Licchesi, J.D.F, Akutsu, M, Komander, D. | | Deposit date: | 2011-06-16 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | An Ankyrin-Repeat Ubiquitin-Binding Domain Determines Trabid'S Specificity for Atypical Ubiquitin Chains.
Nat.Struct.Mol.Biol., 19, 2011
|
|
7K5A
 
 | |
7ST5
 
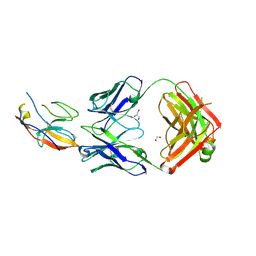 | | Structure of Fab CC-95251 in complex with SIRP-alpha | | Descriptor: | 1,2-ETHANEDIOL, Fab CC-95251 anti-SIRP-alpha heavy chain, Fab CC-95251 anti-SIRP-alpha light chain, ... | | Authors: | Fenalti, G. | | Deposit date: | 2021-11-12 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Preclinical Activity of BMS-986351, an Antibody to SIRP alpha That Enhances Macrophage-mediated Tumor Phagocytosis When Combined with Opsonizing Antibodies.
Cancer Res Commun, 4, 2024
|
|
6DRG
 
 | | NMR solution structure of wild type hFABP1 with GW7647 | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, Fatty acid-binding protein, liver | | Authors: | Scanlon, M.J, Mohanty, B, Doak, B.C, Patil, R. | | Deposit date: | 2018-06-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A ligand-induced structural change in fatty acid-binding protein 1 is associated with potentiation of peroxisome proliferator-activated receptor alpha agonists.
J. Biol. Chem., 294, 2019
|
|
1KU6
 
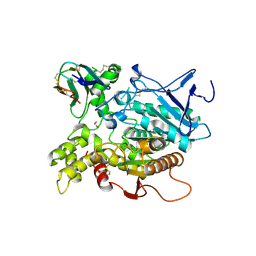 | | Fasciculin 2-Mouse Acetylcholinesterase Complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Bourne, Y, Burmeister, W, Taylor, P, Marchot, P. | | Deposit date: | 2002-01-21 | | Release date: | 2003-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into ligand interactions at the acetylcholinesterase peripheral anionic site.
EMBO J., 22, 2003
|
|
3LGD
 
 | |
5II5
 
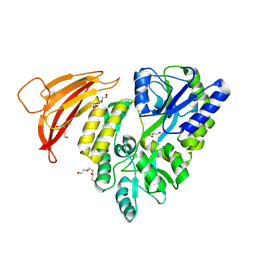 | | Crystal structure of red abalone VERL repeat 1 at 1.8 A resolution | | Descriptor: | Maltose-binding periplasmic protein,Vitelline envelope sperm lysin receptor, TRIETHYLENE GLYCOL, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
3LHQ
 
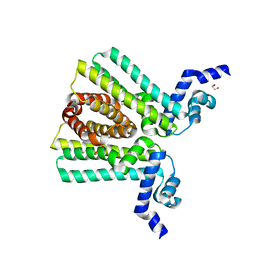 | | DNA-binding transcriptional repressor AcrR from Salmonella typhimurium. | | Descriptor: | 1,2-ETHANEDIOL, AcrAB operon repressor (TetR/AcrR family), DI(HYDROXYETHYL)ETHER | | Authors: | Osipiuk, J, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | X-ray crystal structure of DNA-binding transcriptional repressor AcrR from Salmonella typhimurium.
To be Published
|
|
4BFZ
 
 | |
5Z9C
 
 | |
3LK7
 
 | | The Crystal Structure of UDP-N-acetylmuramoylalanine-D-glutamate (MurD) ligase from Streptococcus agalactiae to 1.5A | | Descriptor: | CHLORIDE ION, SULFATE ION, UDP-N-acetylmuramoylalanine--D-glutamate ligase | | Authors: | Stein, A.J, Sather, A, Shakelford, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-27 | | Release date: | 2010-02-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of UDP-N-acetylmuramoylalanine-D-glutamate (MurD) ligase from Streptococcus agalactiae to 1.5A
To be Published
|
|
4LWU
 
 | | The 1.14A Crystal Structure of Humanized Xenopus MDM2 with RO5499252 | | Descriptor: | (2'S,3R,4'S,5'R)-N-(4-carbamoylphenyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Graves, B.J, Lukacs, C, Janson, C.A. | | Deposit date: | 2013-07-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Discovery of potent and selective spiroindolinone MDM2 inhibitor, RO8994, for cancer therapy.
Bioorg.Med.Chem., 22, 2014
|
|
4E9A
 
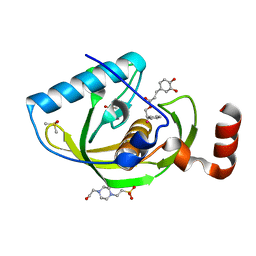 | | Structure of Peptide Deformylase form Helicobacter Pylori in complex with inhibitor | | Descriptor: | 2-phenylethyl (2E)-3-(3,4-dihydroxyphenyl)prop-2-enoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COBALT (II) ION, ... | | Authors: | Cui, K, Zhu, L, Lu, W, Huang, J. | | Deposit date: | 2012-03-20 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | Identification of Novel Peptide Deformylase Inhibitors from Natural Products
To be Published
|
|
9C98
 
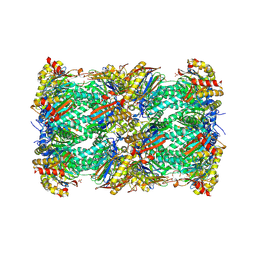 | | Yeast 20S proteasome soaked with isolated TMC-86A | | Descriptor: | MAGNESIUM ION, N-[(2S)-3-hydroxy-1-{[(4S)-1-hydroxy-2-(hydroxymethyl)-6-methyl-3-oxoheptan-4-yl]amino}-1-oxopropan-2-yl]butanamide, Probable proteasome subunit alpha type-7, ... | | Authors: | Meneghello, R, Rustiguel, J.K. | | Deposit date: | 2024-06-13 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | High-throughput protein crystallography to empower natural product-based drug discovery.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
7TEU
 
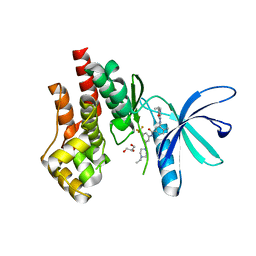 | | Crystal structure of JAK2 JH1 with type II inhibitor YLIU-04-105-1 | | Descriptor: | 3-{(4S)-2-[(cyclopropanecarbonyl)amino]imidazo[1,2-b]pyridazin-6-yl}-N-{3-[(4-ethylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl}-4-methylbenzamide, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Hubbard, S.R. | | Deposit date: | 2022-01-05 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | New scaffolds for type II JAK2 inhibitors overcome the acquired G993A resistance mutation.
Cell Chem Biol, 30, 2023
|
|
5I3W
 
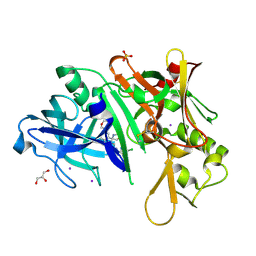 | |
5I46
 
 | | Factor VIIA in complex with the inhibitor (2R,15R)-2-[(1-aminoisoquinolin-6-yl)amino]-8-fluoro-7-hydroxy-4,15,17-trimethyl-13-oxa-4,11-diazatricyclo[14.2.2.1~6,10~]henicosa-1(18),6(21),7,9,16,19-hexaene-3,12-dione | | Descriptor: | (2R,15R)-2-[(1-aminoisoquinolin-6-yl)amino]-8-fluoro-7-hydroxy-4,15,17-trimethyl-13-oxa-4,11-diazatricyclo[14.2.2.1~6,10~]henicosa-1(18),6(21),7,9,16,19-hexaene-3,12-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wei, A. | | Deposit date: | 2016-02-11 | | Release date: | 2016-06-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Atropisomer Control in Macrocyclic Factor VIIa Inhibitors.
J.Med.Chem., 59, 2016
|
|
6D78
 
 | | The complex between high-affinity TCR DMF5(alpha-D26Y,beta-L98W) and human Class I MHC HLA-A2 with the bound MART-1(27-35)peptide | | Descriptor: | Beta-2-microglobulin, DMF5 alpha chain,DMF5 alpha chain, DMF5 beta chain,DMF5 beta chain, ... | | Authors: | Hellman, L.M, Singh, N.K. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.347 Å) | | Cite: | Improving T Cell Receptor On-Target Specificity via Structure-Guided Design.
Mol. Ther., 27, 2019
|
|
4BJ3
 
 | | Integrin alpha2 I domain E318W-collagen complex | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GFOGER PEPTIDE, ... | | Authors: | Carafoli, F, Hamaia, S.W, Bihan, D, Hohenester, E, Farndale, R.W. | | Deposit date: | 2013-04-16 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.042 Å) | | Cite: | An Activating Mutation Reveals a Second Binding Mode of the Integrin Alpha2 I Domain to the Gfoger Motif in Collagens.
Plos One, 8, 2013
|
|
