2HZA
 
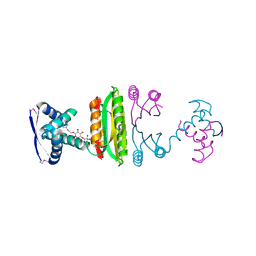 | | Nickel-bound full-length Escherichia coli NikR | | Descriptor: | 3-CYCLOHEXYLPROPYL 4-O-ALPHA-D-GLUCOPYRANOSYL-BETA-D-GLUCOPYRANOSIDE, NICKEL (II) ION, Nickel-responsive regulator | | Authors: | Schreiter, E.R, Drennan, C.L. | | Deposit date: | 2006-08-08 | | Release date: | 2006-08-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | NikR-operator complex structure and the mechanism of repressor activation by metal ions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3BR0
 
 | |
3BQZ
 
 | |
4L9V
 
 | |
5IOY
 
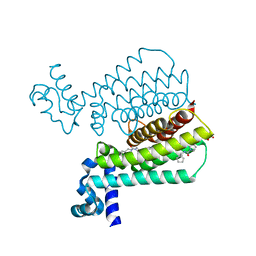 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with N-(cyclopentylmethyl)pyrrolidine-1-carboxamide at 1.77A resolution | | Descriptor: | N-(cyclopentylmethyl)pyrrolidine-1-carboxamide, SULFATE ION, TetR-family transcriptional regulatory repressor protein | | Authors: | Blaszczyk, M, Surade, S, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Fragment-Sized EthR Inhibitors Exhibit Exceptionally Strong Ethionamide Boosting Effect in Whole-Cell Mycobacterium tuberculosis Assays.
ACS Chem. Biol., 12, 2017
|
|
5IPA
 
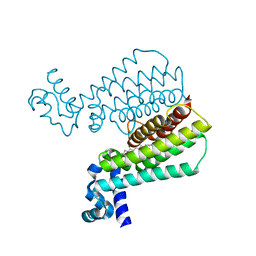 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with (E)-3-(furan-3-yl)-1-(pyrrolidin-1-yl)prop-2-en-1-one at 1.78A resolution | | Descriptor: | (2E)-3-(furan-3-yl)-1-(pyrrolidin-1-yl)prop-2-en-1-one, TetR-family transcriptional regulatory repressor protein | | Authors: | Blaszczyk, M, Surade, S, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Fragment-Sized EthR Inhibitors Exhibit Exceptionally Strong Ethionamide Boosting Effect in Whole-Cell Mycobacterium tuberculosis Assays.
ACS Chem. Biol., 12, 2017
|
|
5IOZ
 
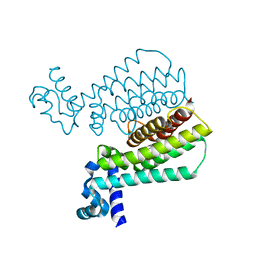 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with N-(cyclopentylmethyl)cyclopentanecarboxamide at 2.02A resolution | | Descriptor: | N-(cyclopentylmethyl)cyclopentanecarboxamide, TetR-family transcriptional regulatory repressor protein | | Authors: | Blaszczyk, M, Surade, S, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Fragment-Sized EthR Inhibitors Exhibit Exceptionally Strong Ethionamide Boosting Effect in Whole-Cell Mycobacterium tuberculosis Assays.
ACS Chem. Biol., 12, 2017
|
|
5IP6
 
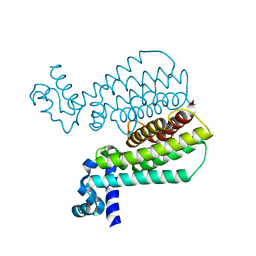 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with N-((tetrahydrofuran-3-yl)methyl)pyrrolidine-1-carboxamide at 1.93A resolution | | Descriptor: | N-{[(3S)-oxolan-3-yl]methyl}pyrrolidine-1-carboxamide, TetR-family transcriptional regulatory repressor protein | | Authors: | Blaszczyk, M, Surade, S, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Fragment-Sized EthR Inhibitors Exhibit Exceptionally Strong Ethionamide Boosting Effect in Whole-Cell Mycobacterium tuberculosis Assays.
ACS Chem. Biol., 12, 2017
|
|
1MYL
 
 | |
4L9J
 
 | |
1QTG
 
 | | AVERAGED NMR MODEL OF SWITCH ARC, A DOUBLE MUTANT OF ARC REPRESSOR | | Descriptor: | Transcriptional repressor arc | | Authors: | Cordes, M.H.J, Walsh, N.P, McKnight, C.J, Sauer, R.T. | | Deposit date: | 1999-06-27 | | Release date: | 1999-07-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Evolution of a protein fold in vitro.
Science, 284, 1999
|
|
1GXR
 
 | | WD40 Region of Human Groucho/TLE1 | | Descriptor: | CALCIUM ION, TRANSDUCIN-LIKE ENHANCER PROTEIN 1 | | Authors: | Pearl, L.H, Roe, S.M, Pickles, L.M. | | Deposit date: | 2002-04-10 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the C-Terminal Wd40 Repeat Domain of the Human Groucho/Tle1 Transcriptional Corepressor
Structure, 10, 2002
|
|
1XGK
 
 | | CRYSTAL STRUCTURE OF N12G AND A18G MUTANT NMRA | | Descriptor: | CHLORIDE ION, GLYCEROL, NITROGEN METABOLITE REPRESSION REGULATOR NMRA, ... | | Authors: | Lamb, H.K, Ren, J, Park, A, Johnson, C, Leslie, K, Cocklin, S, Thompson, P, Mee, C, Cooper, A, Stammers, D.K, Hawkins, A.R. | | Deposit date: | 2004-09-17 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Modulation of the ligand binding properties of the transcription repressor NmrA by GATA-containing DNA and site-directed mutagenesis
Protein Sci., 13, 2004
|
|
2Z4J
 
 | |
3DEU
 
 | | Crystal structure of transcription regulatory protein slyA from Salmonella typhimurium in complex with salicylate ligands | | Descriptor: | 2-HYDROXYBENZOIC ACID, Transcriptional regulator slyA | | Authors: | Le Trong, I, Brzovic, P.S, Fang, F.C, Libby, S.J, Stenkamp, R.E. | | Deposit date: | 2008-06-10 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Evolution of SlyA/RovA Transcription Factors from Repressors to Countersilencers in Enterobacteriaceae .
Mbio, 10, 2019
|
|
3BPX
 
 | | Crystal Structure of MarR | | Descriptor: | 2-HYDROXYBENZOIC ACID, SODIUM ION, Transcriptional regulator | | Authors: | Saridakis, V, Shahinas, D, Xu, X, Christendat, D. | | Deposit date: | 2007-12-19 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insight on the mechanism of regulation of the MarR family of proteins: high-resolution crystal structure of a transcriptional repressor from Methanobacterium thermoautotrophicum.
J.Mol.Biol., 377, 2008
|
|
5MW6
 
 | | Crystal structure of the BCL6 BTB-domain with compound 1 | | Descriptor: | 5-chloranyl-~{N}-(4-chlorophenyl)-2-(3,5-dimethylpyrazol-1-yl)pyrimidin-4-amine, B-cell lymphoma 6 protein | | Authors: | Davies, D.R, Kessler, D. | | Deposit date: | 2017-01-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6.
Cell Rep, 20, 2017
|
|
5MWD
 
 | | Crystal structure of the BCL6 BTB-domain with compound 2 | | Descriptor: | 5-[[5-chloranyl-2-(3,5-dimethylpyrazol-1-yl)pyrimidin-4-yl]amino]-1,3-dihydroindol-2-one, B-cell lymphoma 6 protein | | Authors: | Bader, G, Flotzinger, G, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2017-01-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6.
Cell Rep, 20, 2017
|
|
5MW2
 
 | | CRYSTAL STRUCTURE OF BCL-6 BTB-domain with BI-3802 | | Descriptor: | 2-[6-[[5-chloranyl-2-[(3~{S},5~{R})-3,5-dimethylpiperidin-1-yl]pyrimidin-4-yl]amino]-1-methyl-2-oxidanylidene-quinolin-3-yl]oxy-~{N}-methyl-ethanamide, B-cell lymphoma 6 protein | | Authors: | Bader, G, Flotzinger, G, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2017-01-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6.
Cell Rep, 20, 2017
|
|
2MD5
 
 | | Structure of uninhibited ETV6 ETS domain | | Descriptor: | Transcription factor ETV6 | | Authors: | De, S, Mcintosh, L.P, Chan, A.C, Coyne, H.J, Okon, M, Graves, B.J, Murphy, M.E. | | Deposit date: | 2013-08-29 | | Release date: | 2013-12-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Steric Mechanism of Auto-Inhibitory Regulation of Specific and Non-Specific DNA Binding by the ETS Transcriptional Repressor ETV6.
J.Mol.Biol., 426, 2014
|
|
4IVN
 
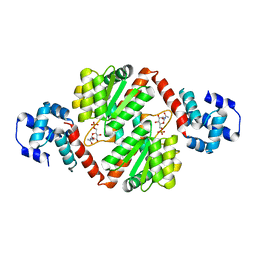 | | The Vibrio vulnificus NanR protein complexed with ManNAc-6P | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-mannopyranose, Transcriptional regulator | | Authors: | Hwang, J, Kim, M.H. | | Deposit date: | 2013-01-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the regulation of sialic acid catabolism by the Vibrio vulnificus transcriptional repressor NanR
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2Q0O
 
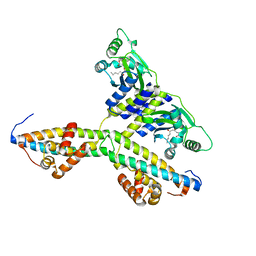 | | Crystal structure of an anti-activation complex in bacterial quorum sensing | | Descriptor: | 3-OXO-OCTANOIC ACID (2-OXO-TETRAHYDRO-FURAN-3-YL)-AMIDE, Probable transcriptional activator protein traR, Probable transcriptional repressor traM | | Authors: | Chen, G, Jeffrey, P.D, Fuqua, C, Shi, Y, Chen, L. | | Deposit date: | 2007-05-22 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for antiactivation in bacterial quorum sensing.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
5NQS
 
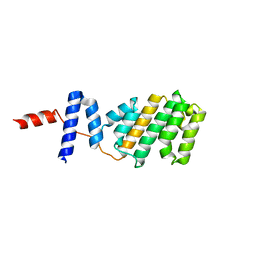 | | Structure of the Arabidopsis Thaliana TOPLESS N-terminal domain | | Descriptor: | Protein TOPLESS | | Authors: | Nanao, M.H, Arevalillo, M.R, Vinos-Poyo, T, Parcy, F, Dumas, R. | | Deposit date: | 2017-04-21 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of the Arabidopsis TOPLESS corepressor provides insight into the evolution of transcriptional repression.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5NQV
 
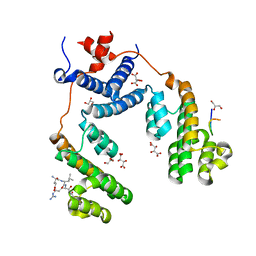 | | Structure of the Arabidopsis Thaliana TOPLESS N-terminal domain | | Descriptor: | EAR motif of IAA27, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Nanao, M.H, Arevalillo, M.R, Vinos-Poyo, T, Parcy, F, Dumas, R. | | Deposit date: | 2017-04-21 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Arabidopsis TOPLESS corepressor provides insight into the evolution of transcriptional repression.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4MGR
 
 | | The crystal structure of Bacillus subtilis GabR, an autorepressor and PLP- and GABA-dependent transcriptional activator of gabT | | Descriptor: | ACETATE ION, HTH-type transcriptional regulatory protein GabR, IMIDAZOLE, ... | | Authors: | Wu, R, Edayathumangalam, R, Garcia, R, Wang, Y, Wang, W, Kreinbring, C.A, Bach, A, Liao, J, Stone, T, Terwilliger, T, Hoang, Q.Q, Belitsky, B.R, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2013-08-28 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
