1L9U
 
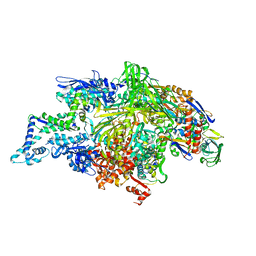 | | THERMUS AQUATICUS RNA POLYMERASE HOLOENZYME AT 4 A RESOLUTION | | Descriptor: | MAGNESIUM ION, RNA POLYMERASE, ALPHA SUBUNIT, ... | | Authors: | Murakami, K.S, Masuda, S, Darst, S.A. | | Deposit date: | 2002-03-26 | | Release date: | 2002-05-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution.
Science, 296, 2002
|
|
8QUE
 
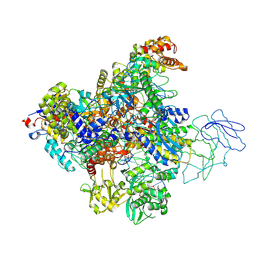 | | Structure of the Bacteriophage PhiKZ non-virion RNA Polymerase bound to DNA and RNA | | Descriptor: | DNA, DNA-directed RNA polymerase, PHIKZ055, ... | | Authors: | de Martin Garrido, N, Yakunina, M, Aylett, C.H.S. | | Deposit date: | 2023-10-16 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the Bacteriophage PhiKZ Non-virion RNA Polymerase Transcribing from its Promoter p119L.
J.Mol.Biol., 436, 2024
|
|
3DXJ
 
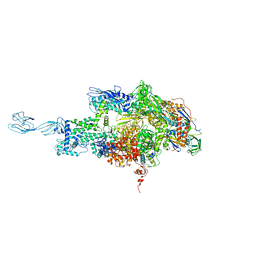 | | Crystal structure of thermus thermophilus rna polymerase holoenzyme in complex with the antibiotic myxopyronin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BACTERIAL RNA POLYMERASE BETA SUBUNIT; CHAIN C, M, ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2008-07-24 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The RNA polymerase "switch region" is a target for inhibitors.
Cell(Cambridge,Mass.), 135, 2008
|
|
1L9Z
 
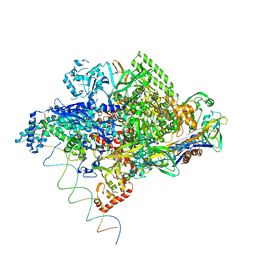 | | Thermus aquaticus RNA Polymerase Holoenzyme/Fork-Junction Promoter DNA Complex at 6.5 A Resolution | | Descriptor: | MAGNESIUM ION, RNA POLYMERASE, ALPHA SUBUNIT, ... | | Authors: | Murakami, K.S, Masuda, S, Campbell, E.A, Muzzin, O, Darst, S.A. | | Deposit date: | 2002-03-27 | | Release date: | 2002-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex.
Science, 296, 2002
|
|
3EQL
 
 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with antibiotic myxopyronin | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Vassylyev, D.G, Vassylyeva, M.N, Artsimovitch, I. | | Deposit date: | 2008-09-30 | | Release date: | 2008-10-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Transcription inactivation through local refolding of the RNA polymerase structure.
Nature, 457, 2009
|
|
2PA8
 
 | |
1MSW
 
 | |
4GWQ
 
 | | Structure of the Mediator Head Module from S. cerevisiae in complex with the carboxy-terminal domain (CTD) of RNA Polymerase II Rpb1 subunit | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, Mediator of RNA polymerase II transcription subunit 11, Mediator of RNA polymerase II transcription subunit 17, ... | | Authors: | Robinson, P.J.J, Bushnell, D.A, Trnka, M.J, Burlingame, A.L, Kornberg, R.D. | | Deposit date: | 2012-09-03 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structure of the Mediator Head module bound to the carboxy-terminal domain of RNA polymerase II.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1GO3
 
 | | Structure of an archeal homolog of the eukaryotic RNA polymerase II RPB4/RPB7 complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE SUBUNIT E, DNA-DIRECTED RNA POLYMERASE SUBUNIT F | | Authors: | Todone, F, Brick, P, Werner, F, Weinzierl, R.O.J, Onesti, S. | | Deposit date: | 2001-10-17 | | Release date: | 2001-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of an Archaeal Homolog of the Eukaryotic RNA Polymerase II Rpb4/Rpb7 Complex
Mol.Cell, 8, 2001
|
|
2RF4
 
 | |
2LO6
 
 | | Structure of Nrd1 CID bound to phosphorylated RNAP II CTD | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, Protein NRD1 | | Authors: | Kubicek, K, Cerna, H, Pasulka, J, Holub, P, Hrossova, D, Loehr, F, Hofr, C, Vanacova, S, Stefl, R. | | Deposit date: | 2012-01-17 | | Release date: | 2012-12-26 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Serine phosphorylation and proline isomerization in RNAP II CTD control recruitment of Nrd1.
Genes Dev., 26, 2012
|
|
1S77
 
 | | T7 RNAP product pyrophosphate elongation complex | | Descriptor: | DNA (5'-D(*GP*CP*CP*GP*TP*GP*CP*GP*CP*AP*TP*TP*CP*GP*CP*CP*GP*TP*GP*TP*T)-3'), DNA (5'-D(*TP*TP*TP*AP*CP*GP*TP*TP*GP*CP*GP*CP*AP*CP*GP*GP*C)-3'), DNA-directed RNA polymerase, ... | | Authors: | Yin, Y.W, Steitz, T.A. | | Deposit date: | 2004-01-29 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | The structural mechanism of translocation and helicase activity in T7 RNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
9BVT
 
 | | RNA Pol II - High Mn(+2) concentration | | Descriptor: | DNA (5'-D(P*AP*CP*GP*TP*CP*CP*CP*TP*CP*TP*CP*GP*A)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Calero, G. | | Deposit date: | 2024-05-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis of transcription: RNA polymerase II substrate binding and metal coordination using a free-electron laser.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7ZWD
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U5 snRNA promoter (CC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZX7
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (CC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZX8
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (OC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
2PI4
 
 | | T7RNAP complexed with a phi10 protein and initiating GTPs. | | Descriptor: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, 5'-D(*CP*TP*TP*CP*CP*TP*AP*TP*AP*GP*TP*GP*AP*GP*TP*CP*GP*TP*AP*TP*TP*A)-3', 5'-D(*TP*AP*AP*TP*AP*CP*GP*AP*CP*TP*CP*AP*CP*T)-3', ... | | Authors: | Kennedy, W.P, Momand, J.R, Yin, Y.W. | | Deposit date: | 2007-04-12 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for de novo RNA synthesis and initiating nucleotide specificity by t7 RNA polymerase.
J.Mol.Biol., 370, 2007
|
|
7SZK
 
 | | Cryo-EM structure of 27a bound to E. coli RNAP and rrnBP1 promoter complex | | Descriptor: | (2S,7R,7aR,13aP,16Z,18E,20S,21S,22R,23R,24R,25S,26R,27S,28E)-5,21,23-trihydroxy-27-methoxy-2,4,16,20,22,24,26-heptamethyl-10-[4-(2-methylpropyl)piperazin-1-yl]-12-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1,6,15-trioxo-1,2,7,7a-tetrahydro-6H-2,7-(epoxypentadeca[1,11,13]trienoimino)[1]benzofuro[4,5-a]phenoxazin-25-yl acetate, DNA (5'-D(P*CP*TP*CP*GP*TP*AP*GP*AP*GP*TP*CP*CP*GP*TP*GP*TP*CP*A)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Shin, Y, Murakami, K.S. | | Deposit date: | 2021-11-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Optimization of Benzoxazinorifamycins to Improve Mycobacterium tuberculosis RNA Polymerase Inhibition and Treatment of Tuberculosis.
Acs Infect Dis., 8, 2022
|
|
7SZJ
 
 | |
8HSR
 
 | | Thermus thermophilus Rho-engaged RNAP elongation complex (composite structure) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (31-MER), ... | | Authors: | Murayama, Y, Ehara, H, Sekine, S. | | Deposit date: | 2022-12-20 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of the transcription termination factor Rho engagement with transcribing RNA polymerase from Thermus thermophilus.
Sci Adv, 9, 2023
|
|
8SYI
 
 | | Cyanobacterial RNAP-EC | | Descriptor: | DNA (37-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Qayyum, M.Z, Imashimizu, M, Leanca, M, Vishwakarma, R.K, Bradley Riaz, A, Yuzenkova, Y, Murakami, K.S. | | Deposit date: | 2023-05-25 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structure and function of the Si3 insertion integrated into the trigger loop/helix of cyanobacterial RNA polymerase.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6VVY
 
 | | Mycobacterium tuberculosis WT RNAP transcription open promoter complex with Sorangicin | | Descriptor: | DNA (65-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Lilic, M, Boyaci, H, Chen, J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VVZ
 
 | | Mycobacterium tuberculosis RNAP S456L mutant transcription initiation intermediate structure with Sorangicin | | Descriptor: | DNA (62-MER), DNA (65-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lilic, M, Boyaci, H, Chen, J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VW0
 
 | | Mycobacterium tuberculosis RNAP S456L mutant open promoter complex | | Descriptor: | DNA (65-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Lilic, M, Boyaci, H, Chen, J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VVX
 
 | | Mycobacterium tuberculosis WT RNAP transcription initiation intermediate structure with Sorangicin | | Descriptor: | DNA (63-MER), DNA (65-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lilic, M, Boyaci, H, Chen, J, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
