3JB9
 
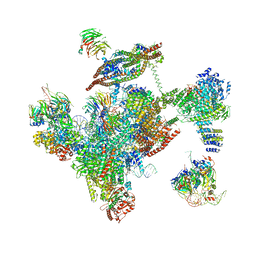 | | Cryo-EM structure of the yeast spliceosome at 3.6 angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yan, C, Hang, J, Wan, R, Huang, M, Wong, C, Shi, Y. | | Deposit date: | 2015-08-09 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a yeast spliceosome at 3.6-angstrom resolution
Science, 349, 2015
|
|
6FT6
 
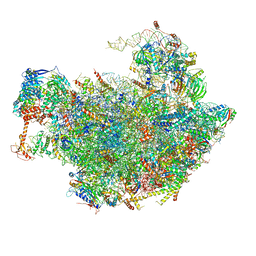 | | Structure of the Nop53 pre-60S particle bound to the exosome nuclear cofactors | | Descriptor: | 25S ribosomal RNA, 5S ribosomal RNA, 60S ribosomal protein L11-A, ... | | Authors: | Schuller, J.M, Falk, S, Conti, E. | | Deposit date: | 2018-02-20 | | Release date: | 2018-03-28 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the nuclear exosome captured on a maturing preribosome.
Science, 360, 2018
|
|
3LKD
 
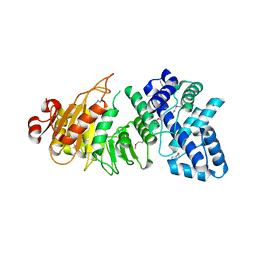 | | Crystal Structure of the type I restriction-modification system methyltransferase subunit from Streptococcus thermophilus, Northeast Structural Genomics Consortium Target SuR80 | | Descriptor: | Type I restriction-modification system methyltransferase subunit | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Mao, M, Xiao, R, Foote, E.L, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-27 | | Release date: | 2010-02-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Northeast Structural Genomics Consortium Target SuR80
To be published
|
|
1XXH
 
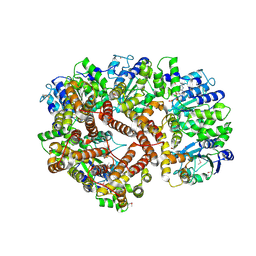 | | ATPgS Bound E. Coli Clamp Loader Complex | | Descriptor: | DNA polymerase III subunit gamma, DNA polymerase III, delta prime subunit, ... | | Authors: | Kazmirski, S.L, Podobnik, M, Weitze, T.F, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2004-11-05 | | Release date: | 2004-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural analysis of the inactive state of the Escherichia coli DNA polymerase clamp-loader complex
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3HKZ
 
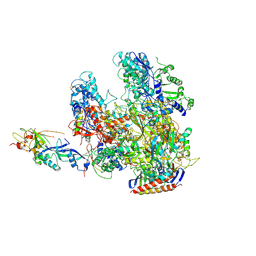 | | The X-ray crystal structure of RNA polymerase from Archaea | | Descriptor: | DNA-directed RNA polymerase subunit 13, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Murakami, K.S. | | Deposit date: | 2009-05-26 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The X-ray crystal structure of RNA polymerase from Archaea.
Nature, 451, 2008
|
|
7W5A
 
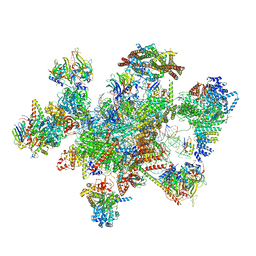 | | The cryo-EM structure of human pre-C*-II complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2021-11-29 | | Release date: | 2022-06-22 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of exon ligation by human spliceosome.
Mol.Cell, 82, 2022
|
|
7W5B
 
 | | The cryo-EM structure of human C* complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2021-11-29 | | Release date: | 2022-06-22 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mechanism of exon ligation by human spliceosome.
Mol.Cell, 82, 2022
|
|
7Z34
 
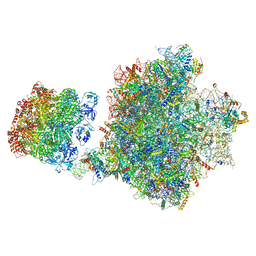 | | Structure of pre-60S particle bound to DRG1(AFG2). | | Descriptor: | 35S pre-ribosomal RNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Prattes, M, Grishkovskaya, I, Bergler, H, Haselbach, D. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-21 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Visualizing maturation factor extraction from the nascent ribosome by the AAA-ATPase Drg1.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6G90
 
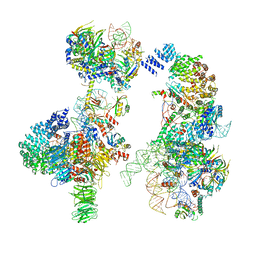 | | Prespliceosome structure provides insight into spliceosome assembly and regulation (map A2) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, Cold sensitive U2 snRNA suppressor 1, Pre-mRNA-processing factor 39, ... | | Authors: | Plaschka, C, Lin, P.-C, Charenton, C, Nagai, K. | | Deposit date: | 2018-04-10 | | Release date: | 2018-08-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Prespliceosome structure provides insights into spliceosome assembly and regulation.
Nature, 559, 2018
|
|
3KHK
 
 | |
7YXM
 
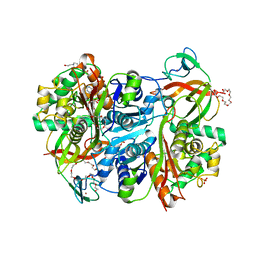 | | Benzoylsuccinyl-CoA thiolase with coenzyme A | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Benzoylsuccinyl-CoA thiolase subunit, ... | | Authors: | Ermler, U, Heider, J, Weidenweber, S. | | Deposit date: | 2022-02-16 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Finis tolueni: a new type of thiolase with an integrated Zn-finger subunit catalyzes the final step of anaerobic toluene metabolism.
Febs J., 289, 2022
|
|
1XXI
 
 | | ADP Bound E. coli Clamp Loader Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase III subunit gamma, DNA polymerase III, ... | | Authors: | Kazmirski, S.L, Podobnik, M, Weitze, T.F, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2004-11-05 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural analysis of the inactive state of the Escherichia coli DNA polymerase clamp-loader complex
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2AR0
 
 | |
1ZJR
 
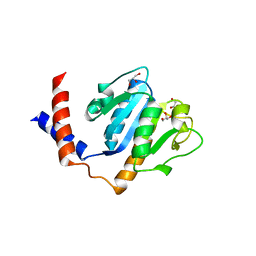 | | Crystal Structure of A. aeolicus TrmH/SpoU tRNA modifying enzyme | | Descriptor: | GLYCEROL, SULFATE ION, tRNA (Guanosine-2'-O-)-methyltransferase | | Authors: | Pleshe, E, Truesdell, J, Batey, R.T. | | Deposit date: | 2005-04-30 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a class II TrmH tRNA-modifying enzyme from Aquifex aeolicus.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
2DFX
 
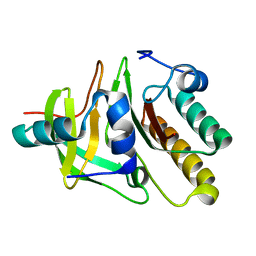 | | Crystal structure of the carboxy terminal domain of colicin E5 complexed with its inhibitor | | Descriptor: | Colicin-E5, Colicin-E5 immunity protein | | Authors: | Yajima, S, Inoue, S, Ogawa, T, Nonaka, T, Ohsawa, K, Masaki, H. | | Deposit date: | 2006-03-06 | | Release date: | 2007-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for sequence-dependent recognition of colicin E5 tRNase by mimicking the mRNA-tRNA interaction
Nucleic Acids Res., 34, 2006
|
|
2DT7
 
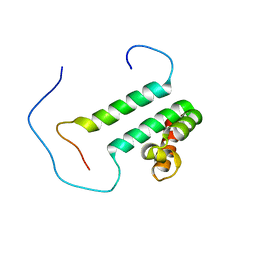 | | Solution structure of the second SURP domain of human splicing factor SF3a120 in complex with a fragment of human splicing factor SF3a60 | | Descriptor: | Splicing factor 3 subunit 1, Splicing factor 3A subunit 3 | | Authors: | He, F, Kuwasako, K, Inoue, M, Guntert, P, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-11 | | Release date: | 2006-12-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the SURP domains and the subunit-assembly mechanism within the splicing factor SF3a complex in 17S U2 snRNP
Structure, 14, 2006
|
|
2DOI
 
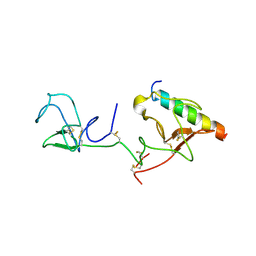 | | The X-ray crystallographic structure of the angiogenesis inhibitor, angiostatin, bound to a peptide from the group A streptococcus protein PAM | | Descriptor: | Angiostatin, Plasminogen-binding group A streptococcal M-like protein PAM | | Authors: | Cnudde, S.E, Prorok, M, Castellino, F.J, Geiger, J.H. | | Deposit date: | 2006-04-29 | | Release date: | 2006-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | X-ray crystallographic structure of the angiogenesis inhibitor, angiostatin, bound to a peptide from the group A streptococcal surface protein PAM
Biochemistry, 45, 2006
|
|
2DOH
 
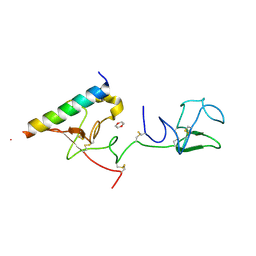 | | The X-ray crystallographic structure of the angiogenesis inhibitor, angiostatin, bound a to a peptide from the group A streptococcal surface protein PAM | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Angiostatin, Plasminogen-binding group A streptococcal M-like protein PAM | | Authors: | Cnudde, S.E, Prorok, M, Castellino, F.J, Geiger, J.H. | | Deposit date: | 2006-04-29 | | Release date: | 2006-12-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystallographic structure of the angiogenesis inhibitor, angiostatin, bound to a peptide from the group A streptococcal surface protein PAM
Biochemistry, 45, 2006
|
|
7ONB
 
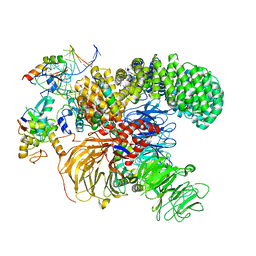 | | Structure of the U2 5' module of the A3'-SSA complex | | Descriptor: | MINX, PHD finger-like domain-containing protein 5A, RNU2, ... | | Authors: | Cretu, C, Pena, V. | | Deposit date: | 2021-05-25 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of intron selection by U2 snRNP in the presence of covalent inhibitors.
Nat Commun, 12, 2021
|
|
7OQE
 
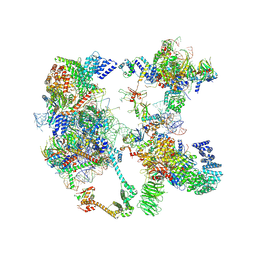 | | Saccharomyces cerevisiae spliceosomal pre-A complex (delta BS-A ACT1) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, ACT1 pre-mRNA (delta BS-A), Cold sensitive U2 snRNA suppressor 1, ... | | Authors: | Zhang, Z, Rigo, N, Dybkov, O, Fourmann, J, Will, C.L, Kumar, V, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2021-06-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural insights into how Prp5 proofreads the pre-mRNA branch site.
Nature, 596, 2021
|
|
7OQB
 
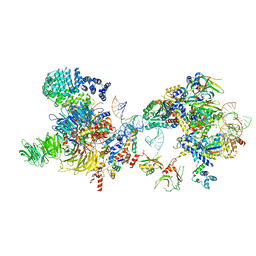 | | The U2 part of Saccharomyces cerevisiae spliceosomal pre-A complex (delta BS-A ACT1) | | Descriptor: | ACT1 pre-mRNA (delta-BS-A), Cold sensitive U2 snRNA suppressor 1, Pre-mRNA-processing ATP-dependent RNA helicase PRP5, ... | | Authors: | Zhang, Z, Rigo, N, Dybkov, O, Fourmann, J, Will, C.L, Kumar, V, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2021-06-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structural insights into how Prp5 proofreads the pre-mRNA branch site.
Nature, 596, 2021
|
|
7F60
 
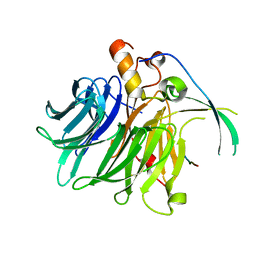 | |
7F90
 
 | |
7OK0
 
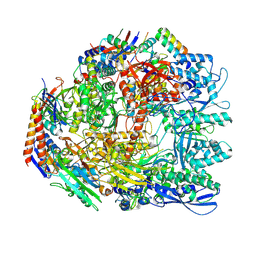 | | Cryo-EM structure of the Sulfolobus acidocaldarius RNA polymerase at 2.88 A | | Descriptor: | Conserved protein, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Pilotto, S, Fouqueau, T, Lukoyanova, N, Sheppard, C, Lucas-Staat, S, Diaz-Santin, L.M, Matelska, D, Prangishvili, D, Cheung, A.C.M, Werner, F. | | Deposit date: | 2021-05-17 | | Release date: | 2021-08-25 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of RNA polymerase inhibition by viral and host factors.
Nat Commun, 12, 2021
|
|
7OQY
 
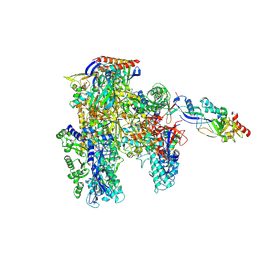 | | Cryo-EM structure of the cellular negative regulator TFS4 bound to the archaeal RNA polymerase | | Descriptor: | Conserved protein, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Pilotto, S, Fouqueau, T, Lukoyanova, N, Sheppard, C, Lucas-Staat, S, Diaz-Santin, L.M, Matelska, D, Prangishvili, D, Cheung, A.C.M, Werner, F. | | Deposit date: | 2021-06-04 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural basis of RNA polymerase inhibition by viral and host factors.
Nat Commun, 12, 2021
|
|
