5JSS
 
 | | Thermolysin in complex with JC149. | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Rational Design of Thermodynamic and Kinetic Binding Profiles by Optimizing Surface Water Networks Coating Protein-Bound Ligands.
J. Med. Chem., 59, 2016
|
|
5JT9
 
 | | Thermolysin in complex with JC106. | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Rational Design of Thermodynamic and Kinetic Binding Profiles by Optimizing Surface Water Networks Coating Protein-Bound Ligands.
J. Med. Chem., 59, 2016
|
|
5JXN
 
 | | Thermolysin in complex with JC240. | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-05-13 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Rational Design of Thermodynamic and Kinetic Binding Profiles by Optimizing Surface Water Networks Coating Protein-Bound Ligands.
J. Med. Chem., 59, 2016
|
|
5JVI
 
 | | Thermolysin in complex with JC148. | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-05-11 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Rational Design of Thermodynamic and Kinetic Binding Profiles by Optimizing Surface Water Networks Coating Protein-Bound Ligands.
J. Med. Chem., 59, 2016
|
|
2VFG
 
 | | Crystal structure of the F96H mutant of Plasmodium falciparum triosephosphate isomerase with 3-phosphoglycerate bound at the dimer interface | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-04 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2VFH
 
 | | Crystal structure of the F96W mutant of Plasmodium falciparum triosephosphate isomerase complexed with 3-phosphoglycerate | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-04 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2P7Z
 
 | | Estrogen Related Receptor Gamma in complex with 4-hydroxy-tamoxifen | | Descriptor: | 4-HYDROXYTAMOXIFEN, Estrogen-related receptor gamma | | Authors: | Abad, M.C. | | Deposit date: | 2007-03-21 | | Release date: | 2008-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural determination of estrogen-related receptor gamma in the presence of phenol derivative compounds.
J.Steroid Biochem.Mol.Biol., 108, 2008
|
|
2VFE
 
 | | Crystal structure of F96S mutant of Plasmodium falciparum triosephosphate isomerase with 3- phosphoglycerate bound at the dimer interface | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, GLYCEROL, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-03 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2VFF
 
 | | Crystal structure of the F96H mutant of Plasmodium falciparum triosephosphate isomerase | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-04 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6VMY
 
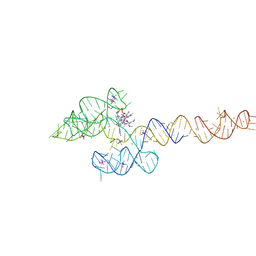 | | Structure of the B. subtilis cobalamin riboswitch | | Descriptor: | Adenosylcobalamin, B. subtilis cobalamin riboswitch, COBALT HEXAMMINE(III), ... | | Authors: | Chan, C.W, Mondragon, A. | | Deposit date: | 2020-01-28 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of an atypical cobalamin riboswitch reveals RNA structural adaptability as basis for promiscuous ligand binding.
Nucleic Acids Res., 48, 2020
|
|
5V7V
 
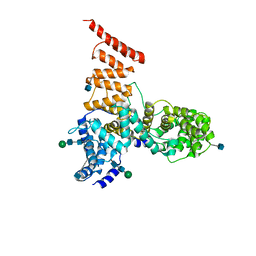 | | Cryo-EM structure of ERAD-associated E3 ubiquitin-protein ligase component HRD3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ERAD-associated E3 ubiquitin-protein ligase component HRD3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Mi, W, Schoebel, S, Stein, A, Rapoport, T.A, Liao, M. | | Deposit date: | 2017-03-20 | | Release date: | 2017-08-16 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3.
Nature, 548, 2017
|
|
8ILR
 
 | | Cryo-EM structure of PI3Kalpha in complex with compound 16 | | Descriptor: | N-[(2S)-1-(ethylamino)-1-oxidanylidene-3-[4-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8IHI
 
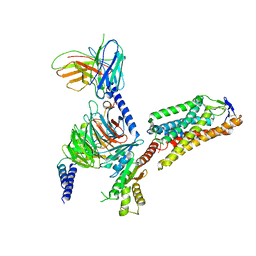 | | Cryo-EM structure of HCA2-Gi complex with acifran | | Descriptor: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHK
 
 | | Cryo-EM structure of HCA3-Gi complex with acifran (local) | | Descriptor: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, Soluble cytochrome b562,Hydroxycarboxylic acid receptor 3 | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHH
 
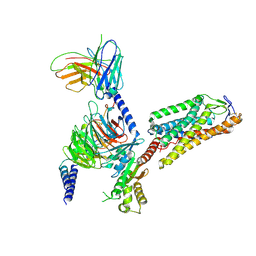 | | Cryo-EM structure of HCA2-Gi complex with LUF6283 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-butyl-1~{H}-pyrazole-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
5UQS
 
 | |
8IHF
 
 | | Cryo-EM structure of HCA2-Gi complex with MK6892 | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHJ
 
 | | Cryo-EM structure of HCA3-Gi complex with acifran | | Descriptor: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8ILV
 
 | | Cryo-EM structure of PI3Kalpha in complex with compound 18 | | Descriptor: | N-[(2R)-1-(ethylamino)-1-oxidanylidene-3-[3-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8IHB
 
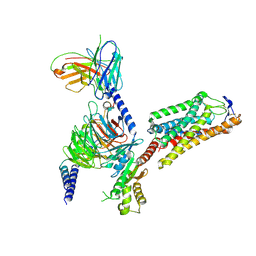 | | Cryo-EM structure of HCA2-Gi complex with GSK256073 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8I7P
 
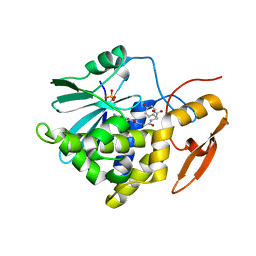 | | Crystal structure of Ricin A chain bound with N2-(2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)glycyl-L-tyrosine | | Descriptor: | 6-(2-ethyl-4-hydroxyphenyl)-1H-indazole-3-carboxamide, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Sakamoto, N, Higashi, S, Kawata, R, Nagatsu, K, Saito, R. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of ricin toxin A chain complexed with a highly potent pterin-based small-molecular inhibitor.
J Enzyme Inhib Med Chem, 38, 2023
|
|
8ILS
 
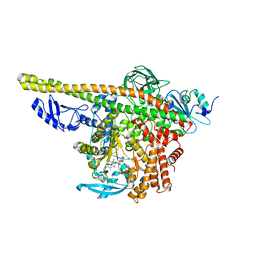 | | Cryo-EM structure of PI3Kalpha in complex with compound 17 | | Descriptor: | N-[(2R)-1-(ethylamino)-1-oxidanylidene-3-[4-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6W3A
 
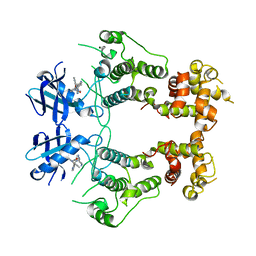 | | Structure of unphosphorylated IRE1 in complex with G-7658 | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1, ~{N}-[6-methyl-5-[3-[2-[[(3~{S})-piperidin-3-yl]amino]pyrimidin-4-yl]pyridin-2-yl]oxy-naphthalen-1-yl]but-2-ynamide | | Authors: | Ferri, E, Wang, W, Joachim, R, Mortara, K. | | Deposit date: | 2020-03-09 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Activation of the IRE1 RNase through remodeling of the kinase front pocket by ATP-competitive ligands.
Nat Commun, 11, 2020
|
|
5UMC
 
 | | Synthesis of novel seleno ureido containing compounds as SLC-0111 analogs. Investigations on carbonic anhydrases activity, glutathione peroxidase and X-ray crystallography | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, UNKNOWN LIGAND, ... | | Authors: | Peat, T.S, Angeli, A, Tanini, D, Bartolucci, G, Capperucci, A, Supuran, C.T, Carta, F. | | Deposit date: | 2017-01-26 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of New Selenoureido Analogues of 4-(4-Fluorophenylureido)benzenesulfonamide as Carbonic Anhydrase Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
7OJR
 
 | |
