6F06
 
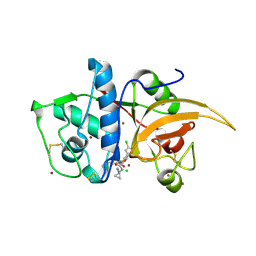 | | CATHEPSIN L IN COMPLEX WITH (3S,14E)-8-(azetidin-3-yl)-19-chloro-N-(1-cyanocyclopropyl)-5-oxo-12,17-dioxa-4-azatricyclo[16.2.2.06,11]docosa-1(21),6,8,10,14,18(22),19-heptaene-3-carboxamide | | Descriptor: | (3~{S},14~{E})-8-(azetidin-3-yl)-19-chloranyl-~{N}-(1-cyanocyclopropyl)-5-oxidanylidene-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(21),6(11),7,9,14,18(22),19-heptaene-3-carboxamide, CHLORIDE ION, Cathepsin L1, ... | | Authors: | Kuglstatter, A, Stihle, M. | | Deposit date: | 2017-11-17 | | Release date: | 2018-04-11 | | Last modified: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
5K8O
 
 | | Mn2+/5NSA-bound 5-nitroanthranilate aminohydrolase | | Descriptor: | 5-nitroanthranilic acid aminohydrolase, 5-nitrosalicylic acid, MANGANESE (II) ION | | Authors: | Kalyoncu, S. | | Deposit date: | 2016-05-30 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Enzymatic hydrolysis by transition-metal-dependent nucleophilic aromatic substitution.
Nat.Chem.Biol., 12, 2016
|
|
6EUM
 
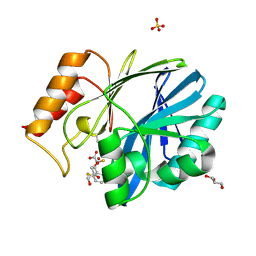 | | CRYSTAL STRUCTURE OF BCII METALLO-BETA-LACTAMASE IN COMPLEX WITH DZ-307 | | Descriptor: | (~{Z})-2-sulfanyl-3-[2,3,6-tris(fluoranyl)phenyl]prop-2-enoic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Stepanovs, D, McDonough, M.A, Schofield, C.J, Zhang, D, El-Husseiny, A, Brem, J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structure activity relationship studies on rhodanines and derived enethiol inhibitors of metallo-beta-lactamases.
Bioorg. Med. Chem., 26, 2018
|
|
5JYK
 
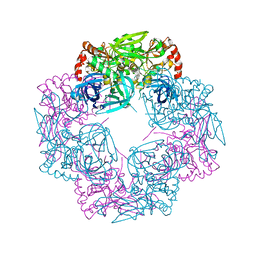 | | Deg9 crystal under 289K | | Descriptor: | GLYCEROL, Protease Do-like 9 | | Authors: | Ouyang, M, Zhang, L.X. | | Deposit date: | 2016-05-14 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | The crystal structure of Deg9 reveals a novel octameric-type HtrA protease
Nat Plants, 3, 2017
|
|
1PW1
 
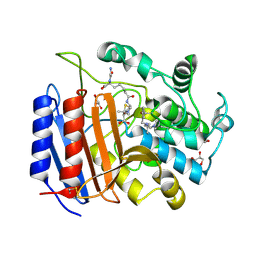 | | Non-Covalent Complex Of Streptomyces R61 DD-Peptidase With A Highly Specific Penicillin | | Descriptor: | (2S,5R,6R)-6-{[(6R)-6-(GLYCYLAMINO)-7-OXIDO-7-OXOHEPTANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLATE, D-alanyl-D-alanine carboxypeptidase, FORMYL GROUP, ... | | Authors: | Silvaggi, N.R, Josephine, H.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2003-06-30 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures of complexes between the R61 DD-peptidase and peptidoglycan-mimetic beta-lactams: a non-covalent complex with a "perfect penicillin"
J.Mol.Biol., 345, 2005
|
|
6T7U
 
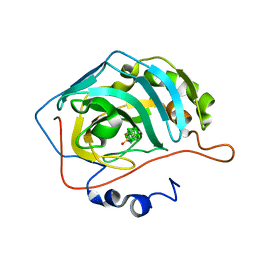 | | Carborane inhibitor of Carbonic Anhydrase IX | | Descriptor: | Carbonic anhydrase 2, Carborane inhibitor, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Kugler, M, Gruner, B. | | Deposit date: | 2019-10-23 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Sulfonamido carboranes as highly selective inhibitors of cancer-specific carbonic anhydrase IX.
Eur.J.Med.Chem., 200, 2020
|
|
5JZE
 
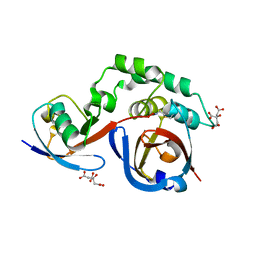 | | Erve virus viral OTU domain protease in complex with mouse ISG15 | | Descriptor: | CITRATE ANION, RNA-dependent RNA polymerase, Ubiquitin-like protein ISG15, ... | | Authors: | Deaton, M.K, Dzimianski, J.V, Pegan, S.D. | | Deposit date: | 2016-05-16 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Biochemical and Structural Insights into the Preference of Nairoviral DeISGylases for Interferon-Stimulated Gene Product 15 Originating from Certain Species.
J.Virol., 90, 2016
|
|
6EW3
 
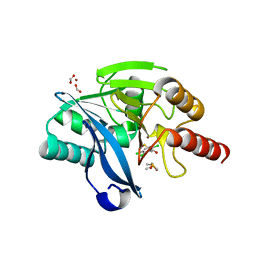 | | Crystal structure of the metallo-beta-lactamase VIM-2 with ML302F | | Descriptor: | (2Z)-2-sulfanyl-3-(2,3,6-trichlorophenyl)prop-2-enoic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Collins, P.M, Brem, J, McDonough, M.A, van Berkel, S.S, von Delft, F, Schofield, C.J. | | Deposit date: | 2017-11-03 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure activity relationship studies on rhodanines and derived enethiol inhibitors of metallo-beta-lactamases.
Bioorg. Med. Chem., 26, 2018
|
|
6EWE
 
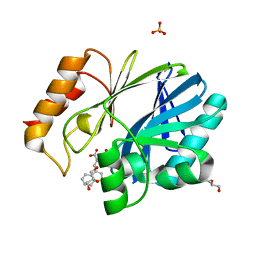 | | Crystal structure of BCII Metallo-beta-lactamase in complex with DZ-308 | | Descriptor: | (~{Z})-3-(1-benzothiophen-3-yl)-2-sulfanyl-prop-2-enoic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Stepanovs, D, McDonough, M.A, Schofield, C.J, Zhang, D, El-Husseiny, A, Brem, J. | | Deposit date: | 2017-11-03 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure activity relationship studies on rhodanines and derived enethiol inhibitors of metallo-beta-lactamases.
Bioorg. Med. Chem., 26, 2018
|
|
6EWK
 
 | | T. californica AChE in complex with a 3-hydroxy-2-pyridine aldoxime. | | Descriptor: | 2-[(~{E})-hydroxyiminomethyl]-6-(5-morpholin-4-ylpentyl)pyridin-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | de la Mora, E, Weik, M, Braiki, A, Mougeot, R, Jean, L, Renard, P.I. | | Deposit date: | 2017-11-04 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Potent 3-Hydroxy-2-Pyridine Aldoxime Reactivators of Organophosphate-Inhibited Cholinesterases with Predicted Blood-Brain Barrier Penetration.
Chemistry, 24, 2018
|
|
5K14
 
 | | HIV-1 Reverse Transcriptase in complex with a 2,6-difluorophenyl DAPY analog | | Descriptor: | 4-{[4-(2,6-difluoro-4-methoxybenzene-1-carbonyl)pyrimidin-2-yl]amino}benzonitrile, HIV-1 reverse transcriptase (isolate LW123), HIV-1 reverse transcriptase(isolate HXB2) | | Authors: | Lansdon, E.B. | | Deposit date: | 2016-05-17 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Novel (2,6-difluorophenyl)(2-(phenylamino)pyrimidin-4-yl)methanones with restricted conformation as potent non-nucleoside reverse transcriptase inhibitors against HIV-1.
Eur.J.Med.Chem., 122, 2016
|
|
6T96
 
 | | Photorhabdus laumondii subsp. laumondii lectin PLL3 | | Descriptor: | Lectin PLL3, SODIUM ION | | Authors: | Melicher, F, Houser, J, Fujdiarova, E, Faltinek, L, Wimmerova, M. | | Deposit date: | 2019-10-26 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Lectin PLL3, a Novel Monomeric Member of the Seven-Bladed beta-Propeller Lectin Family.
Molecules, 24, 2019
|
|
5K8N
 
 | | 5NAA-bound 5-nitroanthranilate aminohydrolase | | Descriptor: | 5-nitroanthranilic acid, 5-nitroanthranilic acid aminohydrolase | | Authors: | Kalyoncu, S. | | Deposit date: | 2016-05-30 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.225 Å) | | Cite: | Enzymatic hydrolysis by transition-metal-dependent nucleophilic aromatic substitution.
Nat.Chem.Biol., 12, 2016
|
|
6T9Z
 
 | | Nidocarborane inhibitor of Carbonic Anhydrase IX | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, Nidocarborane, ... | | Authors: | Brynda, J, Rezacova, P, Kugler, M, Gruner, B. | | Deposit date: | 2019-10-29 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Sulfonamido carboranes as highly selective inhibitors of cancer-specific carbonic anhydrase IX.
Eur.J.Med.Chem., 200, 2020
|
|
6FHG
 
 | |
6TFI
 
 | | PXR IN COMPLEX WITH THROMBIN INHIBITOR COMPOUND 17 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Hillig, R.C, Puetter, V. | | Deposit date: | 2019-11-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design, Synthesis, and Pharmacological Characterization of a Neutral, Non-Prodrug Thrombin Inhibitor with Good Oral Pharmacokinetics.
J.Med.Chem., 63, 2020
|
|
6TGG
 
 | | scFv-1SM3 in complex with glycopeptide containing an sp2-imino sugar | | Descriptor: | 1,2-ETHANEDIOL, Mucin-1, ScFv_SM3, ... | | Authors: | Bermejo, I.A, Navo, C.D, Castro-Lopez, J, Samchez-Fernandez, E.M, Guerreiro, A, Avenoza, A, Busto, J.H, Garcia-Martin, F, Nishimura, S.I, Garcia-Fernandez, J.M, Ortiz-Mellet, C, Bernardes, G.J.L, Hurtado-Guerrero, R, Peregrina, J.M, Corzana, F. | | Deposit date: | 2019-11-15 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis, conformational analysis and in vivo assays of an anti-cancer vaccine that features an unnatural antigen based on an sp 2 -iminosugar fragment.
Chem Sci, 11, 2020
|
|
6F3E
 
 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | 2-[(3~{R})-12-(4-morpholin-4-ylcyclohexyl)oxy-7-thia-9,11-diazatricyclo[6.4.0.0^{2,6}]dodeca-1(8),2(6),9,11-tetraen-3-yl]ethanamide, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Xue, Y, Degorce, S.L, Robb, G.R, Ferguson, A.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Optimization of permeability in a series of pyrrolotriazine inhibitors of IRAK4.
Bioorg. Med. Chem., 26, 2018
|
|
1PWC
 
 | | penicilloyl acyl enzyme complex of the Streptomyces R61 DD-peptidase with penicillin G | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, OPEN FORM - PENICILLIN G | | Authors: | Silvaggi, N.R, Josephine, H.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structures of complexes between the R61 DD-peptidase and peptidoglycan-mimetic beta-lactams: a non-covalent complex with a "perfect penicillin"
J.Mol.Biol., 345, 2005
|
|
6TSN
 
 | | Marasmius oreades agglutinin (MOA), papain back.swap W208Q-Q276W variant | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin, CALCIUM ION, ... | | Authors: | Cordara, G, Manna, D, Krengel, U. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of MOA in complex with a peptide fragment: A protease caught in flagranti .
Curr Res Struct Biol, 2, 2020
|
|
5K8M
 
 | | Apo 5-nitroanthranilate aminohydrolase | | Descriptor: | 5-nitroanthranilic acid aminohydrolase | | Authors: | Kalyoncu, S. | | Deposit date: | 2016-05-30 | | Release date: | 2016-10-05 | | Last modified: | 2016-11-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Enzymatic hydrolysis by transition-metal-dependent nucleophilic aromatic substitution.
Nat.Chem.Biol., 12, 2016
|
|
6FNL
 
 | | Crystal Structure of Ephrin B4 (EphB4) Receptor Protein Kinase | | Descriptor: | Ephrin type-B receptor 4 | | Authors: | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Saxena, K, Schwalbe, H. | | Deposit date: | 2018-02-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.269 Å) | | Cite: | NVP-BHG712: Effects of Regioisomers on the Affinity and Selectivity toward the EPHrin Family.
ChemMedChem, 13, 2018
|
|
5JOP
 
 | | Crystal structure of anti-glycan antibody Fab14.22 in complex with Streptococcus pneumoniae serotype 14 tetrasaccharide at 1.75 A | | Descriptor: | Fab 14.22 light chain, Fab14.22 heavy chain, GLYCEROL, ... | | Authors: | Sarkar, A, Irimia, A, Teyton, L, Wilson, I.A. | | Deposit date: | 2016-05-02 | | Release date: | 2017-03-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | T cells control the generation of nanomolar-affinity anti-glycan antibodies.
J. Clin. Invest., 127, 2017
|
|
5JWP
 
 | | Crystal structure of human FIH D201E variant in complex with Zn, alpha-ketoglutarate, and HIF1 alpha peptide. | | Descriptor: | 2-OXOGLUTARIC ACID, GLYCEROL, Hypoxia-inducible factor 1-alpha, ... | | Authors: | Taabazuing, C.Y, Garman, S.C, Eron, S, Knapp, M.J. | | Deposit date: | 2016-05-12 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The facial triad in the alpha-ketoglutarate dependent oxygenase FIH: A role for sterics in linking substrate binding to O2 activation.
J.Inorg.Biochem., 166, 2016
|
|
1OKZ
 
 | | Structure of human PDK1 kinase domain in complex with UCN-01 | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE 1, 7-HYDROXYSTAUROSPORINE, GLYCEROL, ... | | Authors: | Komander, D, Kular, G.S, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2003-08-01 | | Release date: | 2004-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural Basis for Ucn-01 (7-Hydroxystaurosporine) Specificity and Pdk1 (3-Phosphoinositide-Dependent Protein Kinase-1) Inhibition
Biochem.J., 375, 2003
|
|
