3BIK
 
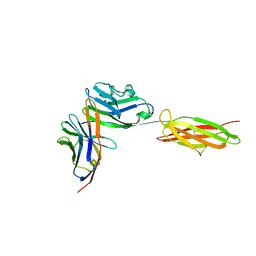 | | Crystal Structure of the PD-1/PD-L1 Complex | | Descriptor: | GLYCEROL, Programmed cell death 1 ligand 1, Programmed cell death protein 1 | | Authors: | Lin, D.Y, Tanaka, Y, Iwasaki, M, Gittis, A.G, Su, H.P, Mikami, B, Okazaki, T, Honjo, T, Minato, N, Garboczi, D.N. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BIS
 
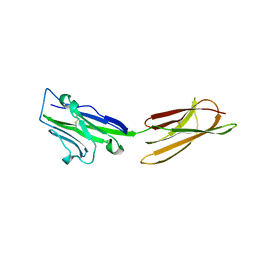 | | Crystal Structure of the PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Lin, D.Y, Tanaka, Y, Iwasaki, M, Gittis, A.G, Su, H.P, Mikami, B, Okazaki, T, Honjo, T, Minato, N, Garboczi, D.N. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5YWO
 
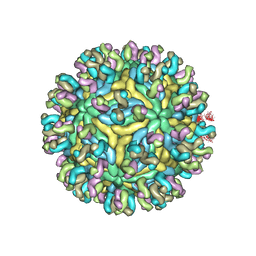 | | Structure of JEV-2F2 Fab complex | | Descriptor: | 2F2 heavy chain, 2F2 light chain, JEV E protein, ... | | Authors: | Qiu, X, Lei, Y.F, Yang, P, Gao, Q, Wang, N, Cao, L, Yuan, S, Wang, X, Xu, Z.K, Rao, Z. | | Deposit date: | 2017-11-29 | | Release date: | 2018-03-21 | | Last modified: | 2018-09-12 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis for neutralization of Japanese encephalitis virus by two potent therapeutic antibodies
Nat Microbiol, 3, 2018
|
|
5YWF
 
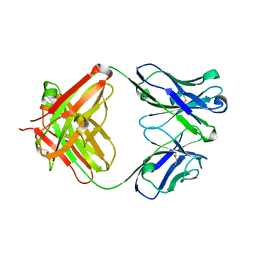 | | Crystal structure of 2H4 Fab | | Descriptor: | 2H4 heavy chain, 2H4 light chain | | Authors: | Qiu, X, Lei, Y, Yang, P, Gao, Q, Wang, N, Cao, L, Wang, X, Xu, Z.K, Rao, Z. | | Deposit date: | 2017-11-29 | | Release date: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structural basis for neutralization of Japanese encephalitis virus by two potent therapeutic antibodies
Nat Microbiol, 3, 2018
|
|
4MUC
 
 | | The 4th and 5th C-terminal domains of Factor H related protein 1 | | Descriptor: | Complement factor H-related protein 1, SULFATE ION | | Authors: | Bhattacharjee, A, Goldman, A, Kolodziejczyk, R, Jokiranta, T.S. | | Deposit date: | 2013-09-21 | | Release date: | 2015-02-18 | | Last modified: | 2015-05-06 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | The Major Autoantibody Epitope on Factor H in Atypical Hemolytic Uremic Syndrome Is Structurally Different from Its Homologous Site in Factor H-related Protein 1, Supporting a Novel Model for Induction of Autoimmunity in This Disease.
J.Biol.Chem., 290, 2015
|
|
2KPY
 
 | | Solution Structure of the major allergen of Artemisia vulgaris (Art v 1) | | Descriptor: | Major pollen allergen Art v 1 | | Authors: | Razzera, G, Gadermaier, G, Almeida, M, Egger, M, Jahn-Schmid, B, Almeida, F, Ferreira, F, Valente, A. | | Deposit date: | 2009-10-23 | | Release date: | 2010-10-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Mapping the interactions between a major pollen allergen and human IgE antibodies.
Structure, 18, 2010
|
|
1WRF
 
 | | Refined solution structure of Der f 2, The Major Mite Allergen from Dermatophagoides farinae | | Descriptor: | Mite group 2 allergen Der f 2 | | Authors: | Ichikawa, S, Takai, T, Inoue, T, Yuuki, T, Okumura, Y, Ogura, K, Inagaki, F, Hatanaka, H. | | Deposit date: | 2004-10-15 | | Release date: | 2005-04-19 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR Study on the Major Mite Allergen Der f 2: Its Refined Tertiary Structure, Epitopes for Monoclonal Antibodies and Characteristics Shared by ML Protein Group Members
J.Biochem.(Tokyo), 137, 2005
|
|
6EUN
 
 | | Crystal structure of Neisseria meningitidis vaccine antigen NadA variant 3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, ... | | Authors: | Dello Iacono, L, Liguori, A, Malito, E, Bottomley, M.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | NadA3 Structures Reveal Undecad Coiled Coils and LOX1 Binding Regions Competed by Meningococcus B Vaccine-Elicited Human Antibodies.
MBio, 9, 2018
|
|
6EUP
 
 | | Crystal structure of Neisseria meningitidis NadA variant 3 double mutant A33I-I38L | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Adhesin A, ... | | Authors: | Dello Iacono, L, Liguori, A, Malito, E, Bottomley, M.J. | | Deposit date: | 2017-10-31 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | NadA3 Structures Reveal Undecad Coiled Coils and LOX1 Binding Regions Competed by Meningococcus B Vaccine-Elicited Human Antibodies.
MBio, 9, 2018
|
|
5YS2
 
 | |
7C81
 
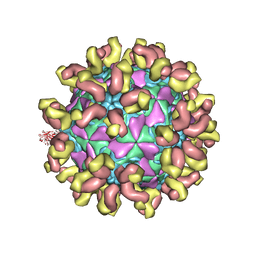 | | E30 F-particle in complex with 6C5 | | Descriptor: | Heavy chain, Light chain, SPHINGOSINE, ... | | Authors: | Wang, K, Zheng, B, Zhang, L, Cui, L, Su, X, Zhang, Q, Guo, Y, Zhu, L, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Serotype specific epitopes identified by neutralizing antibodies underpin immunogenic differences in Enterovirus B.
Nat Commun, 11, 2020
|
|
8JKD
 
 | |
3P30
 
 | | crystal structure of the cluster II Fab 1281 in complex with HIV-1 gp41 ectodomain | | Descriptor: | 1281 Fab heavy chain, 1281 Fab light chain, HIV-1 gp41 | | Authors: | Frey, G, Chen, J, Rits-Volloch, S, Freeman, M.M, Zolla-Pazner, S, Chen, B. | | Deposit date: | 2010-10-04 | | Release date: | 2010-11-17 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Distinct conformational states of HIV-1 gp41 are recognized by neutralizing and non-neutralizing antibodies.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1VER
 
 | | Structure of New Antigen Receptor variable domain from sharks | | Descriptor: | New Antigen Receptor variable domain | | Authors: | Streltsov, V.A. | | Deposit date: | 2004-04-05 | | Release date: | 2004-08-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural evidence for evolution of shark Ig new antigen receptor variable domain antibodies from a cell-surface receptor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
5M15
 
 | | Synthetic nanobody in complex with MBP | | Descriptor: | Maltose-binding periplasmic protein, Synthetic nanobody L2_D09, (a-MBP#3) | | Authors: | Zimmermann, I, Egloff, P, Seeger, M.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-11-15 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthetic single domain antibodies for the conformational trapping of membrane proteins.
Elife, 7, 2018
|
|
5M14
 
 | | Synthetic nanobody in complex with MBP | | Descriptor: | Maltose-binding periplasmic protein, synthetic Nanobody L2_G11 (a-MBP#2) | | Authors: | Zimmermann, I, Egloff, P, Seeger, M.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-11-15 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Synthetic single domain antibodies for the conformational trapping of membrane proteins.
Elife, 7, 2018
|
|
7DNH
 
 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of 2H3 | | Descriptor: | Major capsid protein L1, The heavy chain of 2H3 Fab fragment, The light chain of 2H3 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
7DNK
 
 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of 5G9 | | Descriptor: | Major capsid protein L1, The heavy chain of 5G9 Fab fragment, The light chain of 5G9 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (6.41 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
7DNL
 
 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of A4B4 | | Descriptor: | Major capsid protein L1, The heavy chain of 2H3 Fab fragment, The light chain of A4B4 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
5M13
 
 | | Synthetic nanobody in complex with MBP | | Descriptor: | 1,2-ETHANEDIOL, Maltose-binding periplasmic protein, synthetic Nanobody L2_C06 (a-MBP#2) | | Authors: | Zimmermann, I, Egloff, P, Seeger, M.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-11-15 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.372 Å) | | Cite: | Synthetic single domain antibodies for the conformational trapping of membrane proteins.
Elife, 7, 2018
|
|
6QNA
 
 | | Structure of bovine anti-RSV hybrid Fab B13HC-B4LC | | Descriptor: | B13 Heavy chain, B4 light chain, GLYCEROL | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
6QN9
 
 | | Structure of bovine anti-RSV Fab B4 | | Descriptor: | GLYCEROL, Heavy chain, SULFATE ION, ... | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
6QN7
 
 | | Structure of bovine anti-RSV hybrid Fab B4HC-B13LC | | Descriptor: | Heavy chain of bovine anti-RSV B4, Light chain of bovine anti-RSV B13 | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
3B9K
 
 | | Crystal structure of CD8alpha-beta in complex with YTS 156.7 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab Heavy chain, Fab Light Chain, ... | | Authors: | Shore, D, Wilson, I.A. | | Deposit date: | 2007-11-05 | | Release date: | 2008-11-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of CD8 in Complex with YTS156.7.7 Fab and Interaction with Other CD8 Antibodies Define the Binding Mode of CD8 alphabeta to MHC Class I
J.Mol.Biol., 384, 2008
|
|
5DVM
 
 | | Fc Design 20.8.37 B chain homodimer E357D/S364R/Y407A | | Descriptor: | Fc-III peptide, Ig gamma-1 chain C region | | Authors: | Atwell, S, Leaver-Fay, A, Froning, K.J, Aldaz, H, Pustilnik, A, Lu, F, Huang, F, Yuan, R, Dhanani, S.H, Chamberlain, A.K, Fitchett, J.R, Gutierrez, B, Hendle, J, Demarest, S.J, Kuhlman, B. | | Deposit date: | 2015-09-21 | | Release date: | 2016-03-30 | | Last modified: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Computationally Designed Bispecific Antibodies using Negative State Repertoires.
Structure, 24, 2016
|
|
