7ZPC
 
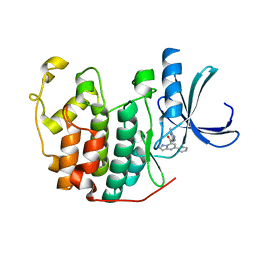 | | CDK2 in complex 9K-DOS | | Descriptor: | Cyclin-dependent kinase 2, ~{N}-(1-methylpyrazol-4-yl)-5-phenyl-pyrazolo[1,5-a]pyrimidine-7-carboxamide | | Authors: | Watt, J.E, Martin, M.P, Noble, M.E.N. | | Deposit date: | 2022-04-27 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Build-Couple-Transform: A Paradigm for Lead-like Library Synthesis with Scaffold Diversity.
J.Med.Chem., 65, 2022
|
|
6FNX
 
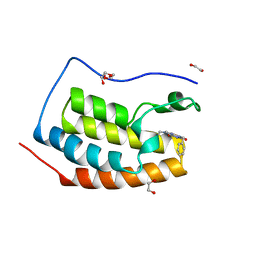 | | FIRST DOMAIN OF HUMAN BROMODOMAIN BRD4 IN COMPLEX WITH INHIBITOR F1 | | Descriptor: | 1,2-ETHANEDIOL, 7-ethyl-3-(phenylmethyl)purine-2,6-dione, Bromodomain-containing protein 4 | | Authors: | Raux, B, Betzi, S. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Integrated Strategy for Lead Optimization Based on Fragment Growing: The Diversity-Oriented-Target-Focused-Synthesis Approach.
J. Med. Chem., 61, 2018
|
|
5ZH2
 
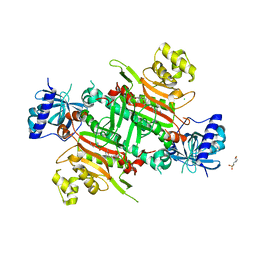 | | CRYSTAL STRUCTURE OF PfKRS WITH INHIBITOR CLADO-5 | | Descriptor: | (3R)-6,8-dihydroxy-3-{[(2R,6R)-6-methyloxan-2-yl]methyl}-3,4-dihydro-1H-2-benzopyran-1-one, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Babbar, P, Malhotra, N, Sharma, M, Harlos, K, Reddy, D.S, Manickam, Y, Sharma, A. | | Deposit date: | 2018-03-11 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Specific Stereoisomeric Conformations Determine the Drug Potency of Cladosporin Scaffold against Malarial Parasite
J. Med. Chem., 61, 2018
|
|
6FR4
 
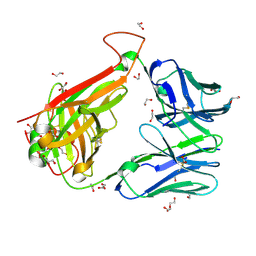 | | 003 TCR Study of CDR Loop Flexibility | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Cole, D.K. | | Deposit date: | 2018-02-15 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | In Silicoand Structural Analyses Demonstrate That Intrinsic Protein Motions Guide T Cell Receptor Complementarity Determining Region Loop Flexibility.
Front Immunol, 9, 2018
|
|
3HKS
 
 | | Crystal structure of eukaryotic translation initiation factor eIF-5A2 from Arabidopsis thaliana | | Descriptor: | 1,2-ETHANEDIOL, Eukaryotic translation initiation factor 5A-2 | | Authors: | Teng, Y.B, He, Y.X, Jiang, Y.L, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2009-05-25 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Arabidopsis translation initiation factor eIF-5A2
Proteins, 77, 2009
|
|
1NPZ
 
 | | Crystal structures of Cathepsin S inhibitor complexes | | Descriptor: | Cathepsin S, N~2~-(morpholin-4-ylcarbonyl)-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-leucinamide | | Authors: | Pauly, T.A, Sulea, T, Ammirati, M, Sivaraman, J, Danley, D.E, Griffor, M.C, Kamath, A.V, Wang, I.K, Laird, E.R, Seddon, A.P, Menard, R, Cygler, M, Rath, V.L. | | Deposit date: | 2003-01-20 | | Release date: | 2003-04-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity determinants of human cathepsin s revealed
by crystal structures of complexes.
Biochemistry, 42, 2003
|
|
6UG3
 
 | | C3 symmetric peptide design number 1, Sporty, crystal form 1 | | Descriptor: | C3-1, Sporty, crystal form 1, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
4YIV
 
 | | Crystal structure of engineered TgAMA1 lacking the DII loop | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Apical membrane antigen AMA1, CADMIUM ION, ... | | Authors: | Parker, M.L, Boulanger, M.J. | | Deposit date: | 2015-03-02 | | Release date: | 2015-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | An Extended Surface Loop on Toxoplasma gondii Apical Membrane Antigen 1 (AMA1) Governs Ligand Binding Selectivity.
Plos One, 10, 2015
|
|
7NDY
 
 | | Di-phosphorylated Barrier-to-Autointegration Factor (BAF) in complex with LEM domain of Emerin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Barrier-to-autointegration factor, N-terminally processed, ... | | Authors: | Marcelot, A, Le Du, M.H, Hoffmann, G, Zinn-Justin, S. | | Deposit date: | 2021-02-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Di-phosphorylated BAF shows altered structural dynamics and binding to DNA, but interacts with its nuclear envelope partners.
Nucleic Acids Res., 49, 2021
|
|
7NNS
 
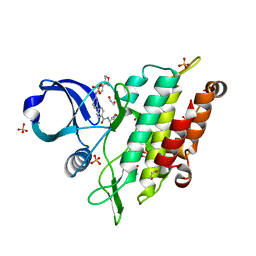 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound Momelotinib | | Descriptor: | 1,2-ETHANEDIOL, Activin receptor type I, Momelotinib, ... | | Authors: | Williams, E, Chen, Z, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound Momelotinib
To Be Published
|
|
4YPQ
 
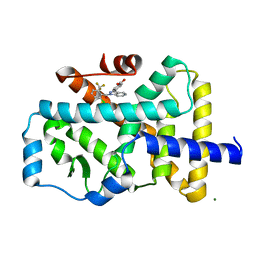 | | Crystal structure of the ROR(gamma)t ligand binding domain in complex with 4-(1-(2-chloro-6-(trifluoromethyl)benzoyl)-1H-indazol-3-yl)benzoic acid | | Descriptor: | 4-{1-[2-chloro-6-(trifluoromethyl)benzoyl]-1H-indazol-3-yl}benzoic acid, MAGNESIUM ION, Nuclear receptor ROR-gamma | | Authors: | Leysen, S, Scheepstra, M, van Almen, G.C, Ottmann, C, Brunsveld, L. | | Deposit date: | 2015-03-13 | | Release date: | 2015-12-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Identification of an allosteric binding site for ROR gamma t inhibition.
Nat Commun, 6, 2015
|
|
1AVA
 
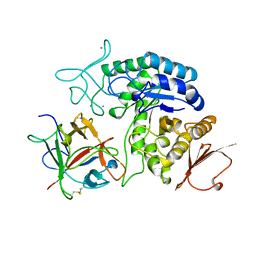 | | AMY2/BASI PROTEIN-PROTEIN COMPLEX FROM BARLEY SEED | | Descriptor: | BARLEY ALPHA-AMYLASE 2(CV MENUET), BARLEY ALPHA-AMYLASE/SUBTILISIN INHIBITOR, CALCIUM ION | | Authors: | Vallee, F, Kadziola, A, Bourne, Y, Juy, M, Svensson, B, Haser, R. | | Deposit date: | 1997-09-15 | | Release date: | 1999-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Barley alpha-amylase bound to its endogenous protein inhibitor BASI: crystal structure of the complex at 1.9 A resolution.
Structure, 6, 1998
|
|
7FZM
 
 | | Crystal Structure of human FABP4 in complex with N-(2,4-dichlorophenyl)-2-(2,6-dihydroxypyrimidin-4-yl)acetamide, i.e. SMILES n1c(cc(nc1O)CC(=O)Nc1c(cc(cc1)Cl)Cl)O with IC50=15 microM | | Descriptor: | Fatty acid-binding protein, adipocyte, N-(2,4-dichlorophenyl)-2-(2,6-dioxo-1,2,5,6-tetrahydropyrimidin-4-yl)acetamide, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Obst-Sander, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
3JR2
 
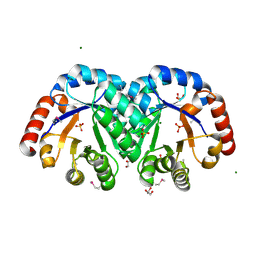 | | X-ray crystal structure of the Mg-bound 3-keto-L-gulonate-6-phosphate decarboxylase from Vibrio cholerae O1 biovar El Tor str. N16961 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Hexulose-6-phosphate synthase SgbH, ... | | Authors: | Nocek, B, Maltseva, N, Stam, J, Anderson, W, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-08 | | Release date: | 2009-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Mg-bound 3-keto-L-gulonate-6-phosphate
decarboxylase from Vibrio cholerae O1 biovar El Tor str. N16961
To be Published
|
|
5AOL
 
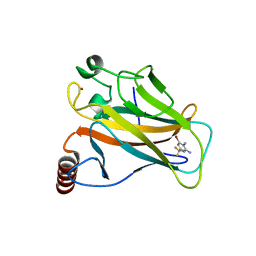 | |
5W6P
 
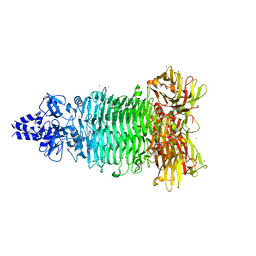 | | Crystal structure of Bacteriophage CBA120 tailspike protein 2 enzymatically active domain (TSP2dN, orf211) | | Descriptor: | 1,2-ETHANEDIOL, POTASSIUM ION, ZINC ION, ... | | Authors: | Plattner, M, Shneider, M.M, Leiman, P.G. | | Deposit date: | 2017-06-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.335 Å) | | Cite: | Structure and Function of the Branched Receptor-Binding Complex of Bacteriophage CBA120.
J.Mol.Biol., 431, 2019
|
|
6UCI
 
 | | Transcription factor DeltaFosB bZIP domain self-assembly, oxidized form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Protein fosB | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2019-09-16 | | Release date: | 2020-01-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Self-assembly of the bZIP transcription factor Delta FosB.
Curr Res Struct Biol, 2, 2020
|
|
4ZSI
 
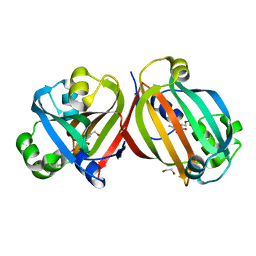 | | Crystal structure of the effector-binding domain of DasR (DasR-EBD) in complex with glucosamine-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, 2-amino-2-deoxy-6-O-phosphono-beta-D-glucopyranose, ... | | Authors: | Fillenberg, S.B, Koerner, S, Muller, Y.A. | | Deposit date: | 2015-05-13 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Crystal Structures of the Global Regulator DasR from Streptomyces coelicolor: Implications for the Allosteric Regulation of GntR/HutC Repressors.
Plos One, 11, 2016
|
|
6LUA
 
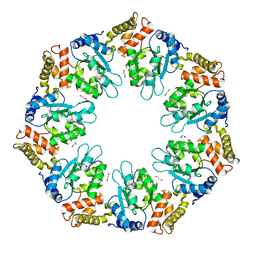 | |
6UHY
 
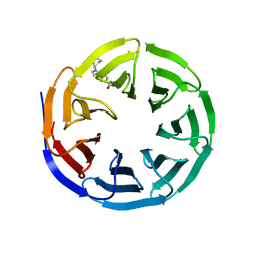 | | WDR5 in complex with Myc site fragment inhibitor | | Descriptor: | 1-cyclohexyl-1H-benzimidazole-5-carboxylic acid, WDR5 | | Authors: | Wang, F, Fesik, S.W. | | Deposit date: | 2019-09-29 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Discovery of WD Repeat-Containing Protein 5 (WDR5)-MYC Inhibitors Using Fragment-Based Methods and Structure-Based Design.
J.Med.Chem., 63, 2020
|
|
7FVV
 
 | | Crystal Structure of human FABP4 in complex with (1S,2R)-2-[(5-carbamoyl-3-ethoxycarbonyl-4-methyl-2-thienyl)carbamoyl]cyclohexanecarboxylic acid | | Descriptor: | (1R,2S)-2-{[5-carbamoyl-3-(ethoxycarbonyl)-4-methylthiophen-2-yl]carbamoyl}cyclohexane-1-carboxylic acid, Fatty acid-binding protein, adipocyte | | Authors: | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
6UNZ
 
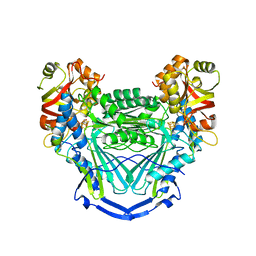 | |
5C3O
 
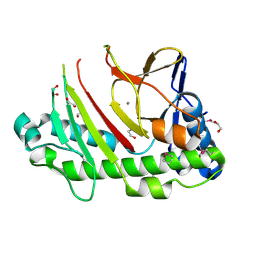 | | Crystal structure of the C-terminal truncated Neurospora crassa T7H (NcT7HdeltaC) in apo form | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Thymine dioxygenase | | Authors: | Li, W, Zhang, T, Ding, J. | | Deposit date: | 2015-06-17 | | Release date: | 2015-10-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the substrate specificity and catalytic mechanism of thymine-7-hydroxylase in fungi
Nucleic Acids Res., 43, 2015
|
|
5C3R
 
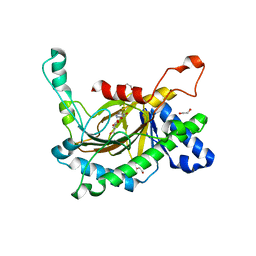 | | Crystal structure of the full-length Neurospora crassa T7H in complex with alpha-KG and 5-hydroxymethyluracil (5hmU) | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, 5-HYDROXYMETHYL URACIL, ... | | Authors: | Li, W, Zhang, T, Ding, J. | | Deposit date: | 2015-06-17 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis for the substrate specificity and catalytic mechanism of thymine-7-hydroxylase in fungi
Nucleic Acids Res., 43, 2015
|
|
5V35
 
 | | Crystal structure of V71F mutant of the FKBP domain of human aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1) complexed with S-farnesyl-L-cysteine methyl ester | | Descriptor: | Aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1), FARNESYL | | Authors: | Yadav, R.P, Gakhar, L, Liping, Y, Artemyev, N.O. | | Deposit date: | 2017-03-06 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unique structural features of the AIPL1-FKBP domain that support prenyl lipid binding and underlie protein malfunction in blindness.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
