7SNN
 
 | | Disulfide stabilized HIV-1 CA hexamer in complex with capsid inhibitor N-(1-(3-(4-chloro-1-methyl-3-(methylsulfonamido)-1H-indazol-7-yl)-4-oxo-3,4-dihydropyrido[2,3-d]pyrimidin-2-yl)-2-(3,5-difluorophenyl)ethyl)-2-(3-(difluoromethyl)-5,6-dihydrocyclopenta[c]pyrazol-1(4H)-yl)acetamide | | Descriptor: | CHLORIDE ION, Capsid protein p24, IODIDE ION, ... | | Authors: | Bester, S.M, Kvaratskhelia, M. | | Deposit date: | 2021-10-28 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Disulfide stabilized HIV-1 CA hexamer in complex with capsid inhibitor N-(1-(3-(4-chloro-1-methyl-3-(methylsulfonamido)-1H-indazol-7-yl)-4-oxo-3,4-dihydropyrido[2,3-d]pyrimidin-2-yl)-2-(3,5-difluorophenyl)ethyl)-2-(3-(difluoromethyl)-5,6-dihydrocyclopenta[c]pyrazol-1(4H)-yl)acetamide
To Be Published
|
|
4EB7
 
 | | A. fulgidus IscS-IscU complex structure | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, ... | | Authors: | Marinoni, E.N, de Oliveira, J.S, Nicolet, Y, Raulfs, E.C, Amara, P, Dean, D.R, Fontecilla-Camps, J.C. | | Deposit date: | 2012-03-23 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | (IscS-IscU)2 complex structures provide insights into Fe2S2 biogenesis and transfer.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
1NZI
 
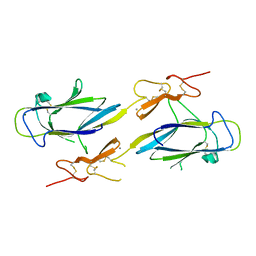 | | Crystal Structure of the CUB1-EGF Interaction Domain of Complement Protease C1s | | Descriptor: | CALCIUM ION, Complement C1s component, MAGNESIUM ION | | Authors: | Gregory, L.A, Thielens, N.M, Arlaud, G.J, Fontecilla-Camps, J.C, Gaboriaud, C. | | Deposit date: | 2003-02-18 | | Release date: | 2003-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray structure of the Ca2+-binding interaction domain of C1s. Insights into the assembly of the C1 complex of complement
J.Biol.Chem., 278, 2003
|
|
8CF2
 
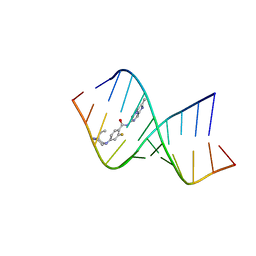 | | Solution structure of the RNA helix formed by the 5'-end of U1 snRNA and an A-1 bulged 5'-splice site in complex with SMN-CY | | Descriptor: | 4-[(3~{S})-3-ethylpiperazin-1-yl]-2-fluoranyl-~{N}-(2-methylimidazo[1,2-a]pyrazin-6-yl)benzamide, RNA (5'-R(P*AP*UP*AP*CP*(PSU)P*(PSU)P*AP*CP*CP*UP*G)-3'), RNA (5'-R(P*GP*GP*AP*GP*UP*AP*AP*GP*UP*CP*U)-3') | | Authors: | Malard, F, Marquevielle, J, Campagne, S. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The diversity of splicing modifiers acting on A-1 bulged 5'-splice sites reveals rules for rational drug design.
Nucleic Acids Res., 52, 2024
|
|
8JC7
 
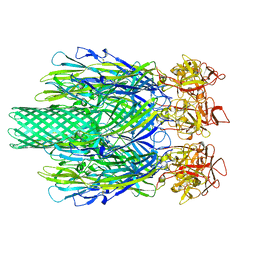 | | Cryo-EM structure of Vibrio campbellii alpha-hemolysin | | Descriptor: | CALCIUM ION, Hemolysin, POTASSIUM ION | | Authors: | Wang, C.H, Yeh, M.K, Ho, M.C, Lin, S.M. | | Deposit date: | 2023-05-10 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.06 Å) | | Cite: | Structural basis for calcium-stimulating pore formation of Vibrio alpha-hemolysin.
Nat Commun, 14, 2023
|
|
1QOJ
 
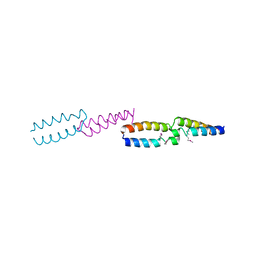 | | Crystal Structure of E.coli UvrB C-terminal domain, and a model for UvrB-UvrC interaction. | | Descriptor: | UVRB | | Authors: | Sohi, M, Alexandrovich, A, Moolenaar, G, Visse, R, Goosen, N, Vernede, X, Fontecilla-Camps, J, Champness, J, Sanderson, M.R. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-10 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of E.Coli Uvrb C-Terminal Domain, and a Model for Uvrb-Uvrc Interaction
FEBS Lett., 465, 2000
|
|
1QDD
 
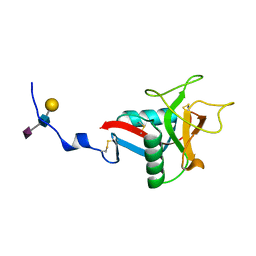 | | CRYSTAL STRUCTURE OF HUMAN LITHOSTATHINE TO 1.3 A RESOLUTION | | Descriptor: | LITHOSTATHINE, beta-D-galactopyranose-(1-3)-[N-acetyl-alpha-neuraminic acid-(2-6)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Gerbaud, V, Pignol, D, Loret, E, Bertrand, J.A, Berland, Y, Fontecilla-Camps, J.C, Canselier, J.P, Gabas, N, Verdier, J.M. | | Deposit date: | 1999-05-20 | | Release date: | 1999-05-28 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of calcite crystal growth inhibition by the N-terminal undecapeptide of lithostathine.
J.Biol.Chem., 275, 2000
|
|
1T2B
 
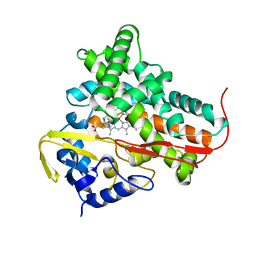 | | Crystal Structure of cytochrome P450cin complexed with its substrate 1,8-cineole | | Descriptor: | 1,3,3-TRIMETHYL-2-OXABICYCLO[2.2.2]OCTANE, P450cin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Meharenna, Y.T, Li, H, Hawkes, D.B, Pearson, A.G, De Voss, J, Poulos, T.L. | | Deposit date: | 2004-04-20 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of P450cin in a complex with its substrate, 1,8-cineole, a close structural homologue to D-camphor, the substrate for P450cam
Biochemistry, 43, 2004
|
|
2VOS
 
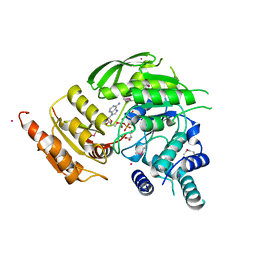 | | Mycobacterium tuberculosis Folylpolyglutamate synthase complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, COBALT (II) ION, FOLYLPOLYGLUTAMATE SYNTHASE PROTEIN FOLC, ... | | Authors: | Young, P.G, Baker, E.N, Metcalf, P, Smith, C.A. | | Deposit date: | 2008-02-19 | | Release date: | 2008-07-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Mycobacterium Tuberculosisfolylpolyglutamate Synthase Complexed with Adp and Amppcp.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1T6L
 
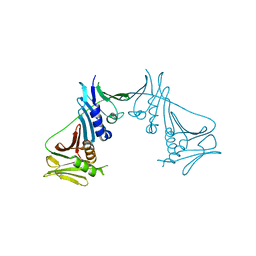 | | Crystal Structure of the Human Cytomegalovirus DNA Polymerase Subunit, UL44 | | Descriptor: | DNA polymerase processivity factor | | Authors: | Appleton, B.A, Loregian, A, Filman, D.J, Coen, D.M, Hogle, J.M. | | Deposit date: | 2004-05-06 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Cytomegalovirus DNA Polymerase Subunit UL44 Forms a C Clamp-Shaped Dimer.
Mol.Cell, 15, 2004
|
|
1A37
 
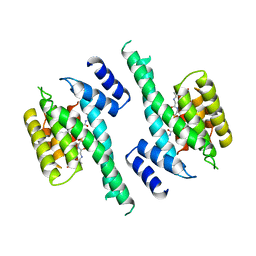 | | 14-3-3 PROTEIN ZETA BOUND TO PS-RAF259 PEPTIDE | | Descriptor: | 14-3-3 PROTEIN ZETA, PS-RAF259 PEPTIDE LSQRQRST(SEP)TPNVHM | | Authors: | Petosa, C, Masters, S.C, Pohl, J, Wang, B, Fu, H, Liddington, R.C. | | Deposit date: | 1998-01-28 | | Release date: | 1999-03-02 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | 14-3-3zeta binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove.
J.Biol.Chem., 273, 1998
|
|
1BC0
 
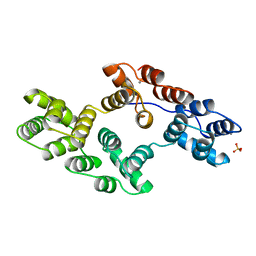 | | RECOMBINANT RAT ANNEXIN V, W185A MUTANT | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Swairjo, M.A, Li, C.W, Head, J.F, Seaton, B.A. | | Deposit date: | 1998-05-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational and crystallographic analyses of interfacial residues in annexin V suggest direct interactions with phospholipid membrane components.
Biochemistry, 37, 1998
|
|
4F3H
 
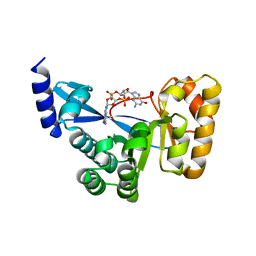 | | The structural of FimXEAL-c-di-GMP from Xanthomonas campestris | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Putative uncharacterized protein | | Authors: | Chin, K.-H, Liao, Y.-T, Chou, S.-H. | | Deposit date: | 2012-05-09 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural polymorphism of c-di-GMP bound to an EAL domain and in complex with a type II PilZ-domain protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4F48
 
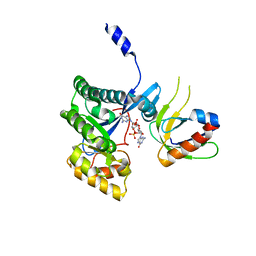 | | The X-ray structural of FimXEAL-c-di-GMP-PilZ complexes from Xanthomonas campestris | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Putative uncharacterized protein, Type IV fimbriae assembly protein | | Authors: | Chin, K.-H, Liao, Y.-T, Chou, S.-H. | | Deposit date: | 2012-05-10 | | Release date: | 2012-11-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural polymorphism of c-di-GMP bound to an EAL domain and in complex with a type II PilZ-domain protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1MSM
 
 | | The HIV protease (mutant Q7K L33I L63I) complexed with KNI-764 (an inhibitor) | | Descriptor: | (4R)-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-N-(2-methylbenzyl)-1,3-thiazolidine-4-carboxamide, POL polyprotein | | Authors: | Vega, S, Kang, L.-W, Velazquez-Campoy, A, Kiso, Y, Amzel, L.M, Freire, E. | | Deposit date: | 2002-09-19 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural and thermodynamic escape mechanism from a drug resistant mutation of the HIV-1 protease.
Proteins, 55, 2004
|
|
1O53
 
 | |
4HVK
 
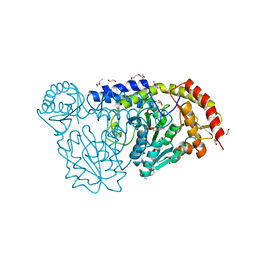 | | Crystal structure and functional studies of an unusual L-cysteine desulfurase from Archaeoglobus fulgidus. | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yamanaka, Y, Zeppieri, L, Nicolet, Y, Marinoni, E.N, de Oliveira, J.S, Masafumi, O, Dean, D.R, Fontecilla-Camps, J.C. | | Deposit date: | 2012-11-06 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure and functional studies of an unusual L-cysteine desulfurase from Archaeoglobus fulgidus.
Dalton Trans, 42, 2013
|
|
1TUM
 
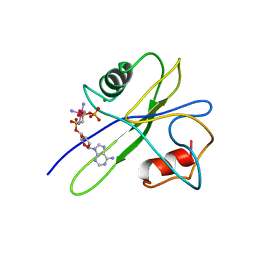 | | MUTT PYROPHOSPHOHYDROLASE-METAL-NUCLEOTIDE-METAL COMPLEX, NMR, 16 STRUCTURES | | Descriptor: | COBALT TETRAAMMINE ION, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, ... | | Authors: | Lin, J, Abeygunawardana, C, Frick, D.N, Bessman, M.J, Mildvan, A.S. | | Deposit date: | 1996-12-05 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the quaternary MutT-M2+-AMPCPP-M2+ complex and mechanism of its pyrophosphohydrolase action.
Biochemistry, 36, 1997
|
|
4I8C
 
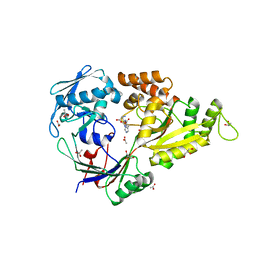 | | X-ray structure of NikA in complex with Ni-(L-His)2 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lebrette, H, Iannello, M, Fontecilla-Camps, J.C, Cavazza, C. | | Deposit date: | 2012-12-03 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | The binding mode of Ni-((L)-His)(2) in NikA revealed by X-ray crystallography.
J.Inorg.Biochem., 121C, 2012
|
|
1LOA
 
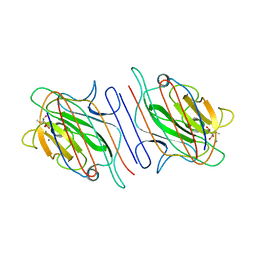 | |
6M54
 
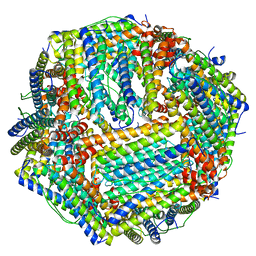 | | Human apo ferritin frozen on TEM grid with Amorphous nickel titanium alloy supporting film | | Descriptor: | FE (II) ION, Ferritin heavy chain | | Authors: | Huang, X, Zhang, L, Wen, Z, Chen, H, Li, S, Ji, G, Yin, C, Sun, F. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Amorphous nickel titanium alloy film: A new choice for cryo electron microscopy sample preparation.
Prog.Biophys.Mol.Biol., 156, 2020
|
|
6APF
 
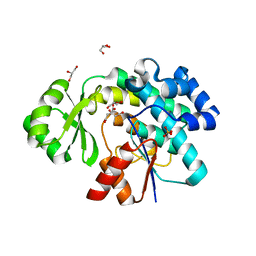 | | Trans-acting transferase from Disorazole synthase complexed with Citrate. | | Descriptor: | CITRIC ACID, DisD protein, GLYCEROL, ... | | Authors: | Mathews, I.I, Lyubimov, A, Soltis, M, Khosla, C, Cohen, A, Robbins, T. | | Deposit date: | 2017-08-17 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The Conformational Flexibility of the Acyltransferase from the Disorazole Polyketide Synthase Is Revealed by an X-ray Free-Electron Laser Using a Room-Temperature Sample Delivery Method for Serial Crystallography.
Biochemistry, 56, 2017
|
|
8D1U
 
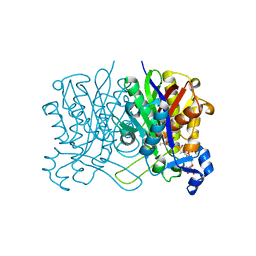 | | E. coli beta-ketoacyl-[acyl carrier protein] synthase III (FabH) with an acetylated cysteine and in complex with oxa(dethia)-Coenzyme A | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3, CHLORIDE ION, oxa(dethia)-CoA | | Authors: | Benjamin, A.B, Stunkard, L.M, Ling, J, Nice, J.N, Lohman, J.R. | | Deposit date: | 2022-05-27 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Structures of chloramphenicol acetyltransferase III and Escherichia coli beta-ketoacylsynthase III co-crystallized with partially hydrolysed acetyl-oxa(dethia)CoA.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
6XLC
 
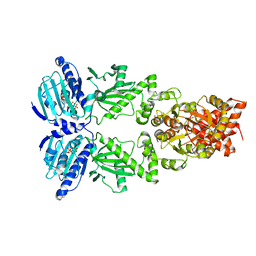 | | Full-length Hsc82 bound to AMPPNP | | Descriptor: | ATP-dependent molecular chaperone HSC82, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Liu, Y.X, Sun, M, Myasnikov, A.G, Elnatan, D, Agard, D.A. | | Deposit date: | 2020-06-28 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Full-length Hsc82 bound to AMPPNP
To Be Published
|
|
6PIS
 
 | |
