1PTX
 
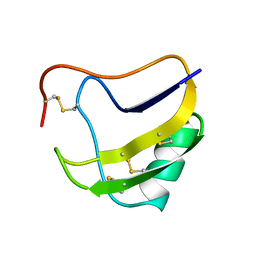 | |
1RCN
 
 | | CRYSTAL STRUCTURE OF THE RIBONUCLEASE A D(APTPAPAPG) COMPLEX : DIRECT EVIDENCE FOR EXTENDED SUBSTRATE RECOGNITION | | Descriptor: | DNA (5'-D(*AP*TP*AP*A)-3'), PROTEIN (RIBONUCLEASE A (E.C.3.1.27.5)) | | Authors: | Fontecilla-Camps, J.C, De Llorens, R, Le Du, M.H, Cuchillo, C.M. | | Deposit date: | 1994-05-27 | | Release date: | 1994-09-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure of ribonuclease A.d(ApTpApApG) complex. Direct evidence for extended substrate recognition.
J.Biol.Chem., 269, 1994
|
|
4URH
 
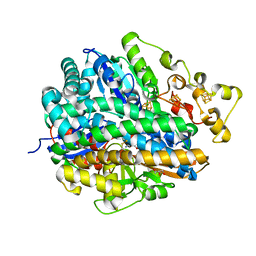 | | High-resolution structure of partially oxidized D. fructosovorans NiFe-hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystallographic studies of [NiFe]-hydrogenase mutants: towards consensus structures for the elusive unready oxidized states.
J. Biol. Inorg. Chem., 20, 2015
|
|
4UQP
 
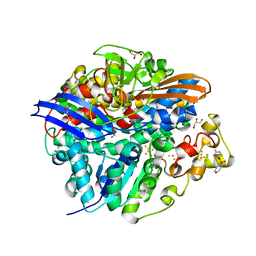 | | High-resolution structure of the D. fructosovorans NiFe-hydrogenase L122A mutant after exposure to air | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-29 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystallographic Studies of [Nife]-Hydrogenase Mutants: Towards Consensus Structures for the Elusive Unready Oxidized States.
J.Biol.Inorg.Chem., 20, 2015
|
|
4UQL
 
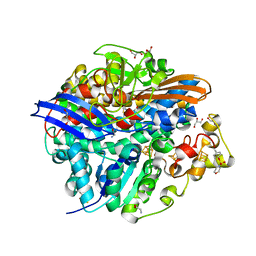 | | High-resolution structure of a Ni-A Ni-Sox mixture of the D. fructosovorans NiFe-hydrogenase L122A mutant | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystallographic Studies of [Nife]-Hydrogenase Mutants: Towards Consensus Structures for the Elusive Unready Oxidized States.
J.Biol.Inorg.Chem., 20, 2015
|
|
1AHO
 
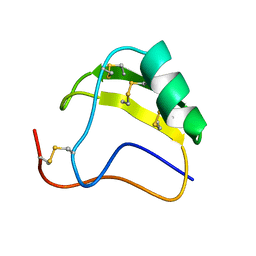 | | THE AB INITIO STRUCTURE DETERMINATION AND REFINEMENT OF A SCORPION PROTEIN TOXIN | | Descriptor: | TOXIN II | | Authors: | Smith, G.D, Blessing, R.H, Ealick, S.E, Fontecilla-Camps, J.C, Hauptman, H.A, Housset, D, Langs, D.A, Miller, R. | | Deposit date: | 1997-04-08 | | Release date: | 1997-10-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Ab initio structure determination and refinement of a scorpion protein toxin.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
8QTO
 
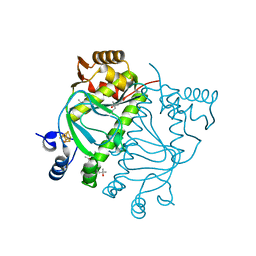 | | CRYSTAL STRUCTURE OF HOLO-L28H-FNR OF A. FISCHERI | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FNR type regulator, IRON/SULFUR CLUSTER | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2023-10-13 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the Reactivity of [4Fe-4S] Fumarate and Nitrate Reduction (FNR) Regulator with O2 and NO: Increased O2 Resistance and Relative Specificity for NO of the [4Fe-4S] L28H FNR Cluster
Inorganics (Basel), 11, 2023
|
|
7B0C
 
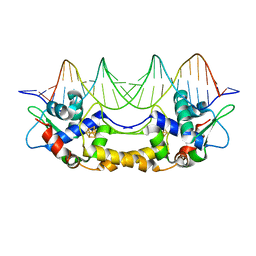 | | [4Fe-4S]-NsrR complexed to 23-bp HmpA1 operator fragment | | Descriptor: | DNA (5'-D(P*AP*AP*CP*AP*CP*GP*AP*AP*TP*AP*TP*CP*AP*TP*CP*TP*AP*CP*CP*AP*AP*TP*T)-3'), DNA (5'-D(P*AP*AP*TP*TP*GP*GP*TP*AP*GP*AP*TP*GP*AP*TP*AP*TP*TP*CP*GP*TP*GP*TP*T)-3'), HTH-type transcriptional repressor NsrR, ... | | Authors: | Rohac, R, Fontecilla-Camps, J.C, Volbeda, A. | | Deposit date: | 2020-11-19 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural determinants of DNA recognition by the NO sensor NsrR and related Rrf2-type [FeS]-transcription factors.
Commun Biol, 5, 2022
|
|
1CC1
 
 | | CRYSTAL STRUCTURE OF A REDUCED, ACTIVE FORM OF THE NI-FE-SE HYDROGENASE FROM DESULFOMICROBIUM BACULATUM | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE (II) ION, HYDROGENASE (LARGE SUBUNIT), ... | | Authors: | Garcin, E, Vernede, X, Hatchikian, E.C, Volbeda, A, Frey, M, Fontecilla-Camps, J.C. | | Deposit date: | 1999-03-03 | | Release date: | 1999-06-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a reduced [NiFeSe] hydrogenase provides an image of the activated catalytic center
Structure Fold.Des., 7, 1999
|
|
6Y45
 
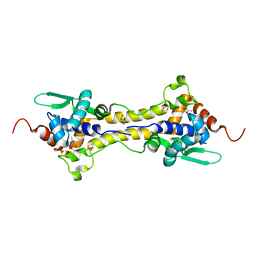 | | Crystal Structure of the H33A variant of RsrR | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Rohac, R, Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Electron and Proton Transfers Modulate DNA Binding by the Transcription Regulator RsrR.
J.Am.Chem.Soc., 142, 2020
|
|
6Y42
 
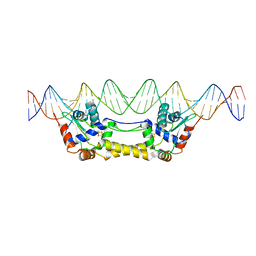 | |
1A27
 
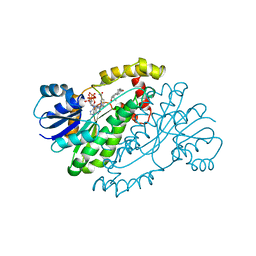 | | HUMAN 17-BETA-HYDROXYSTEROID-DEHYDROGENASE TYPE 1 C-TERMINAL DELETION MUTANT COMPLEXED WITH ESTRADIOL AND NADP+ | | Descriptor: | 17-BETA-HYDROXYSTEROID-DEHYDROGENASE, ESTRADIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mazza, C, Breton, R, Housset, D, Fontecilla-Camps, J.-C. | | Deposit date: | 1998-01-16 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human Type I 17Beta-Hydroxysteroid Dehydrogenase: Site Directed Mutagenesis and X-Ray Crystallography Structure-Function Analysis
Thesis, Universite Joseph Fourier, 1997
|
|
3H3X
 
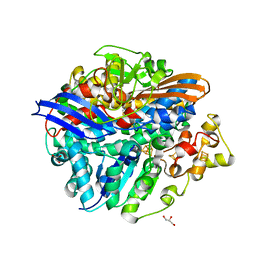 | | Structure of the V74M large subunit mutant of NI-FE hydrogenase in an oxidized state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martinez, N, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2009-04-17 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Introduction of methionines in the gas channel makes [NiFe] hydrogenase aero-tolerant
J.Am.Chem.Soc., 131, 2009
|
|
5LQS
 
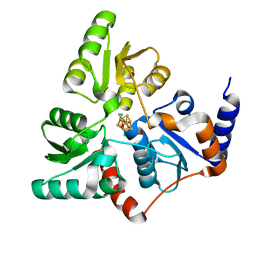 | | Structure of quinolinate synthase Y21F mutant in complex with substrate-derived quinolinate | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, QUINOLINIC ACID, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2016-08-17 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Quinolinate Synthase in Complex with a Substrate Analogue, the Condensation Intermediate, and Substrate-Derived Product.
J.Am.Chem.Soc., 138, 2016
|
|
5LQM
 
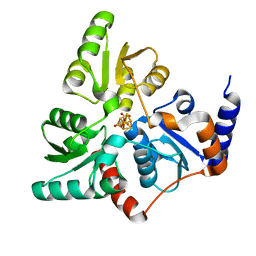 | |
6G94
 
 | |
6F48
 
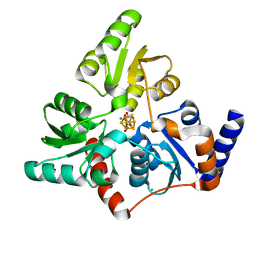 | | Structure of quinolinate synthase with reaction intermediates X and Y | | Descriptor: | 2-imino,3-carboxy,5-hydroxy,6-oxo hexanoic acid, 5-hydroxy,-4,5-dihydroquinolinate, CHLORIDE ION, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic Trapping of Reaction Intermediates in Quinolinic Acid Synthesis by NadA.
ACS Chem. Biol., 13, 2018
|
|
6F4L
 
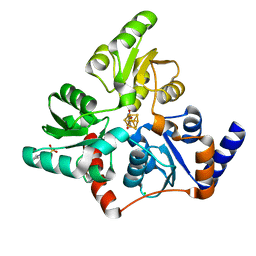 | |
6F4D
 
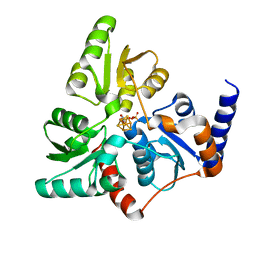 | |
6G74
 
 | |
2Q60
 
 | | Crystal structure of the ligand binding domain of polyandrocarpa misakiensis rxr in tetramer in absence of ligand | | Descriptor: | Retinoid X receptor | | Authors: | Borel, F, De Groot, A, Juillan-Binard, C, De Rosny, E, Laudet, V, Pebay-Peyroula, E, Fontecilla-Camps, J.-C, Ferrer, J.-L. | | Deposit date: | 2007-06-04 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the ligand-binding domain of the retinoid X receptor from the ascidian polyandrocarpa misakiensis.
Proteins, 74, 2008
|
|
4KL8
 
 | | High-resolution structure of anaerobically purified Dm. baculatum [NiFeSe]-hydrogenase after crystallization under air | | Descriptor: | CALCIUM ION, CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, ... | | Authors: | Volbeda, A, Cavazza, C, Fontecilla-Camps, J.C. | | Deposit date: | 2013-05-07 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural foundations for the O2 resistance of Desulfomicrobium baculatum [NiFeSe]-hydrogenase.
Chem.Commun.(Camb.), 49, 2013
|
|
4KO1
 
 | | High X-ray dose structure of H2-activated anaerobically purified Dm. baculatum [NiFeSe]-hydrogenase after crystallization under air | | Descriptor: | CALCIUM ION, CARBONMONOXIDE-(DICYANO) IRON, GLYCEROL, ... | | Authors: | Volbeda, A, Cavazza, C, Fontecilla-Camps, J.C. | | Deposit date: | 2013-05-11 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural foundations for the O2 resistance of Desulfomicrobium baculatum [NiFeSe]-hydrogenase.
Chem.Commun.(Camb.), 49, 2013
|
|
5N07
 
 | |
5N08
 
 | |
