8X7P
 
 | | CCA-bound E46K alpha-synuclein fibrils | | Descriptor: | Alpha-synuclein, copper;trisodium;18-(2-carboxylatoethyl)-20-(carboxylatomethyl)-12-ethenyl-7-ethyl-3,8,13,17-tetramethyl-17,18-dihydroporphyrin-21,23-diide-2-carboxylate | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2023-11-24 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X7O
 
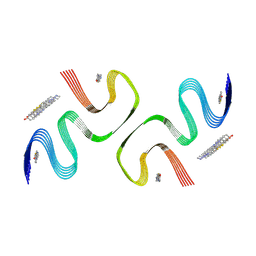 | | PiB-bound E46K mutanted alpha-synuclein fibrils | | Descriptor: | 2-[4-(methylamino)phenyl]-1,3-benzothiazol-6-ol, Alpha-synuclein | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2023-11-24 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X7M
 
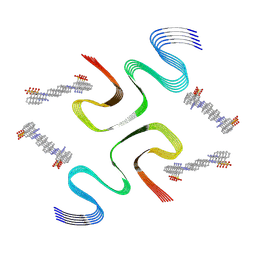 | | CR-bound E46K alpha-synuclein fibrils | | Descriptor: | Alpha-synuclein, sodium 3,3'-(1E,1'E)-biphenyl-4,4'-diylbis(diazene-2,1-diyl)bis(4-aminonaphthalene-1-sulfonate) | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2023-11-24 | | Release date: | 2024-09-11 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X7L
 
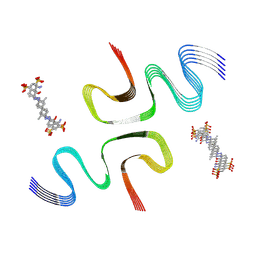 | | EB-bound E46K alpha-synuclein fibrils | | Descriptor: | 4-azanyl-6-[[4-[4-[(~{E})-(8-azanyl-1-oxidanyl-5,7-disulfo-naphthalen-2-yl)diazenyl]-3-methyl-phenyl]-2-methyl-phenyl]diazenyl]-5-oxidanyl-naphthalene-1,3-disulfonic acid, Alpha-synuclein | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2023-11-24 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7EQH
 
 | |
8X7B
 
 | | ThT-bound E46K alpha-synuclein fibrils | | Descriptor: | 2-[4-(dimethylamino)phenyl]-3,6-dimethyl-1,3-benzothiazol-3-ium, Alpha-synuclein | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2023-11-23 | | Release date: | 2024-11-27 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8ZLI
 
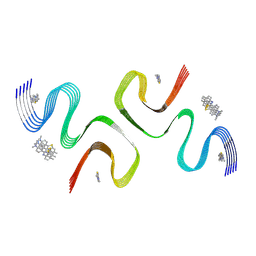 | | BTA-2-bound E46K alpha-synuclein fibrils | | Descriptor: | Alpha-synuclein, ~{N},~{N}-dimethyl-4-(6-methyl-1,3-benzothiazol-2-yl)aniline | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2024-05-20 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8ZLO
 
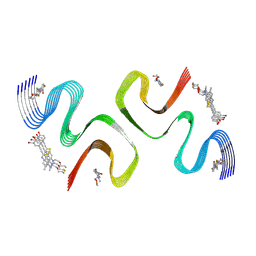 | | F0502B-bound E46K alpha-synuclein fibril | | Descriptor: | 2-bromanyl-4-[(~{E})-2-[6-[2-(2-fluoranylethoxy)ethyl-methyl-amino]-5-methyl-1,3-benzothiazol-2-yl]ethenyl]phenol, Alpha-synuclein | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2024-05-20 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3SDK
 
 | | Structure of yeast 20S open-gate proteasome with Compound 34 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-[(2S)-3-(3-tert-butyl-1,2,4-oxadiazol-5-yl)-1-({(2S)-1-[(4-methylbenzyl)amino]-1-oxo-4-phenylbutan-2-yl}amino)-1-oxopropan-2-yl]-5-methyl-1,2-oxazole-3-carboxamide, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2011-06-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Optimization of a series of dipeptides with a P3 threonine residue as non-covalent inhibitors of the chymotrypsin-like activity of the human 20S proteasome.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3SDI
 
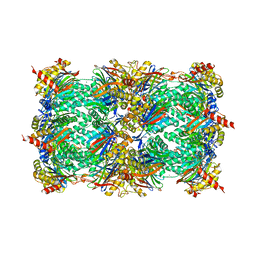 | | Structure of yeast 20S open-gate proteasome with Compound 20 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N~4~-(2,2-dimethylpropyl)-N~1~-{(2S)-1-[(4-methylbenzyl)amino]-1-oxo-4-phenylbutan-2-yl}-N~2~-[(5-methyl-1,2-oxazol-3-yl)carbonyl]-L-aspartamide, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2011-06-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Optimization of a series of dipeptides with a P3 threonine residue as non-covalent inhibitors of the chymotrypsin-like activity of the human 20S proteasome.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
8R1S
 
 | |
8QVQ
 
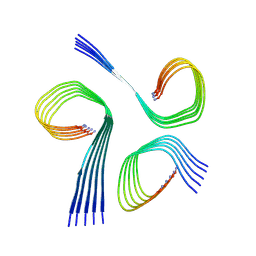 | |
8QJ1
 
 | |
8QVR
 
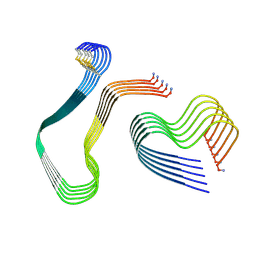 | |
8OHI
 
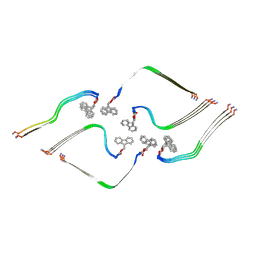 | | Structure of the Fmoc-Tau-PAM4 Type 2 amyloid fibril | | Descriptor: | Microtubule-associated protein tau | | Authors: | Wilkinson, M, Louros, N, Tsaka, G, Ramakers, M, Morelli, C, Garcia, T, Gallardo, R.U, D'Haeyer, S, Goossens, V, Audenaert, D, Thal, D.R, Ranson, N.A, Radford, S.E, Rousseau, F, Schymkowitz, J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Local structural preferences in shaping tau amyloid polymorphism.
Nat Commun, 15, 2024
|
|
8OH2
 
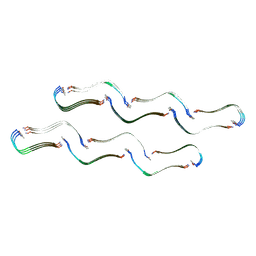 | | Structure of the Tau-PAM4 Type 1 amyloid fibril | | Descriptor: | Microtubule-associated protein tau | | Authors: | Wilkinson, M, Louros, N, Tsaka, G, Ramakers, M, Morelli, C, Garcia, T, Gallardo, R.U, D'Haeyer, S, Goossens, V, Audenaert, D, Thal, D.R, Ranson, N.A, Radford, S.E, Rousseau, F, Schymkowitz, J. | | Deposit date: | 2023-03-20 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Local structural preferences in shaping tau amyloid polymorphism.
Nat Commun, 15, 2024
|
|
8OHP
 
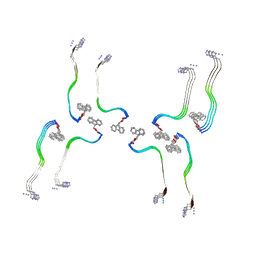 | | Structure of the Fmoc-Tau-PAM4 Type 3 amyloid fibril | | Descriptor: | Microtubule-associated protein tau | | Authors: | Wilkinson, M, Louros, N, Tsaka, G, Ramakers, M, Morelli, C, Garcia, T, Gallardo, R.U, D'Haeyer, S, Goossens, V, Audenaert, D, Thal, D.R, Ranson, N.A, Radford, S.E, Rousseau, F, Schymkowitz, J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Local structural preferences in shaping tau amyloid polymorphism.
Nat Commun, 15, 2024
|
|
8OI0
 
 | | Structure of the Fmoc-Tau-PAM4 Type 4 amyloid fibril | | Descriptor: | Microtubule-associated protein tau | | Authors: | Wilkinson, M, Louros, N, Tsaka, G, Ramakers, M, Morelli, C, Garcia, T, Gallardo, R.U, D'Haeyer, S, Goossens, V, Audenaert, D, Thal, D.R, Ranson, N.A, Radford, S.E, Rousseau, F, Schymkowitz, J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Local structural preferences in shaping tau amyloid polymorphism.
Nat Commun, 15, 2024
|
|
5UE4
 
 | | proMMP-9desFnII complexed to JNJ0966 INHIBITOR | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-9, SULFATE ION, ... | | Authors: | Alexander, R.S, Spurlino, J, Milligan, C. | | Deposit date: | 2016-12-29 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a highly selective chemical inhibitor of matrix metalloproteinase-9 (MMP-9) that allosterically inhibits zymogen activation.
J. Biol. Chem., 292, 2017
|
|
5JDD
 
 | |
5JDJ
 
 | | Crystal structure of domain I10 from titin in space group P212121 | | Descriptor: | CALCIUM ION, Titin | | Authors: | Williams, R, Bogomolovas, J, Labiet, S, Mayans, O. | | Deposit date: | 2016-04-16 | | Release date: | 2016-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | Exploration of pathomechanisms triggered by a single-nucleotide polymorphism in titin's I-band: the cardiomyopathy-linked mutation T2580I.
Open Biology, 6, 2016
|
|
2IC4
 
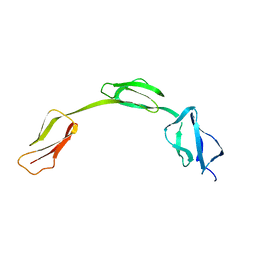 | | Solution structure of the His402 allotype of the Factor H SCR6-SCR7-SCR8 fragment | | Descriptor: | Complement factor H | | Authors: | Fernando, A.N, Furtado, P.B, Gilbert, H.E, Clark, S.J, Day, A.J, Sim, R.B, Perkins, S.J. | | Deposit date: | 2006-09-12 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Associative and Structural Properties of the Region of Complement Factor H Encompassing the Tyr402His Disease-related Polymorphism and its Interactions with Heparin.
J.Mol.Biol., 368, 2007
|
|
6EJK
 
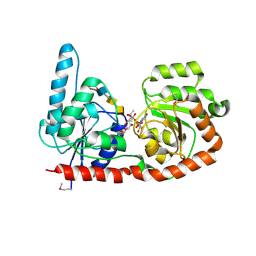 | | Structure of a glycosyltransferase | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-glucopyranose, NerylNeryl pyrophosphate, Uridine-Diphosphate-Methylene-N-acetyl-galactosamine, ... | | Authors: | Ramirez, A.S, Boilevin, J, Mehdipour, A.R, Hummer, G, Darbre, T, Reymond, J.L, Locher, K.P. | | Deposit date: | 2017-09-21 | | Release date: | 2018-02-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of the molecular ruler mechanism of a bacterial glycosyltransferase.
Nat Commun, 9, 2018
|
|
8AZ3
 
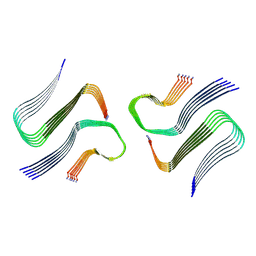 | | IAPP S20G growth-phase fibril polymorph 4PF-CU | | Descriptor: | Islet amyloid polypeptide | | Authors: | Wilkinson, M, Xu, Y, Gallardo, R, Radford, S.E, Ranson, N.A. | | Deposit date: | 2022-09-05 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural evolution of fibril polymorphs during amyloid assembly.
Cell, 186, 2023
|
|
8AZ7
 
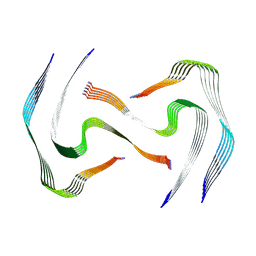 | | IAPP S20G plateau-phase fibril polymorph 4PF-LJ | | Descriptor: | Islet amyloid polypeptide | | Authors: | Wilkinson, M, Xu, Y, Gallardo, R, Radford, S.E, Ranson, N.A. | | Deposit date: | 2022-09-05 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural evolution of fibril polymorphs during amyloid assembly.
Cell, 186, 2023
|
|
