5F6X
 
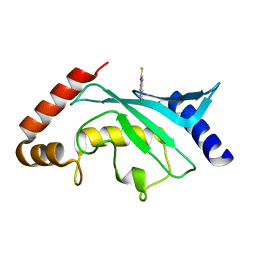 | | Crystal structure of Ubc9 (K48/K49A/E54A) complexed with Fragment 2 (mercaptobenzoxazole from cocktail screen) | | Descriptor: | 5-chloranyl-3~{H}-1,3-benzoxazole-2-thione, SUMO-conjugating enzyme UBC9 | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5F6E
 
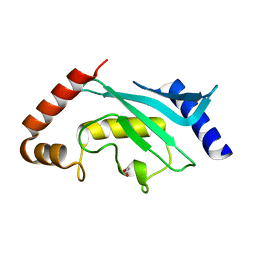 | | Crystal Structure of human Ubc9 (K48A/K49A/E54A) | | Descriptor: | 1,2-ETHANEDIOL, SUMO-conjugating enzyme UBC9 | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, Z, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-05 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5F6U
 
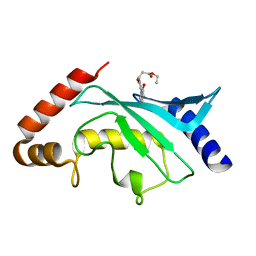 | | Crystal Structure of Ubc9 (K48A/K49A/E54A) complexed with Fragment 8 (JSS190B146) | | Descriptor: | SUMO-conjugating enzyme UBC9, ethyl 3-[4-(2-hydroxyphenyl)-3-oxidanyl-phenyl]propanoate | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, Z, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5F6Y
 
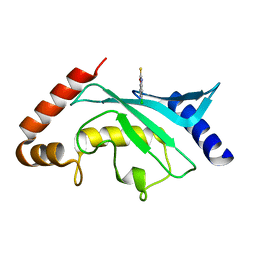 | | Crystal structure of Ubc9 (K48/K49A/E54A) complexed with Fragment 2 (mercaptobenzoxazole) | | Descriptor: | 5-chloranyl-3~{H}-1,3-benzoxazole-2-thione, SUMO-conjugating enzyme UBC9 | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
7Z88
 
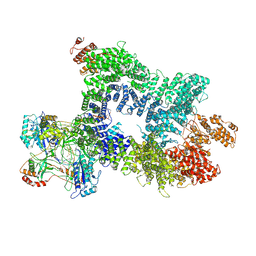 | | DNA-PK in the intermediate state | | Descriptor: | (~{S})-[2-chloranyl-4-fluoranyl-5-(7-morpholin-4-ylquinazolin-4-yl)phenyl]-(6-methoxypyridazin-3-yl)methanol, DNA (26-MER), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Liang, S, Blundell, T.L. | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Human DNA-dependent protein kinase activation mechanism.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Z87
 
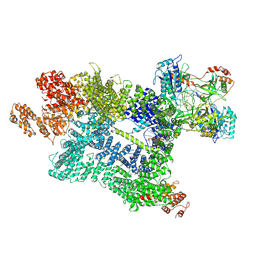 | | DNA-PK in the active state | | Descriptor: | (~{S})-[2-chloranyl-4-fluoranyl-5-(7-morpholin-4-ylquinazolin-4-yl)phenyl]-(6-methoxypyridazin-3-yl)methanol, DNA (26-MER), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Liang, S, Blundell, T.L. | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Human DNA-dependent protein kinase activation mechanism.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3BV4
 
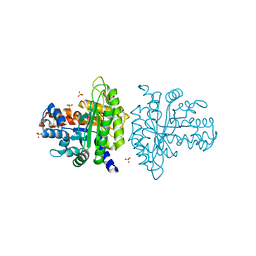 | | Crystal structure of a rabbit muscle fructose-1,6-bisphosphate aldolase A dimer variant | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase A, SULFATE ION | | Authors: | Sherawat, M, Tolan, D.R, Allen, K.N. | | Deposit date: | 2008-01-04 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a rabbit muscle fructose-1,6-bisphosphate aldolase A dimer variant.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3BQO
 
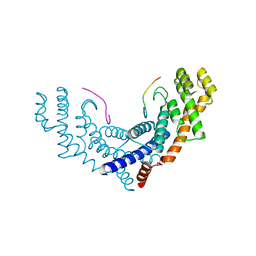 | | Crystal Structure of TRF1 TRFH domain and TIN2 peptide complex | | Descriptor: | TERF1-interacting nuclear factor 2, Telomeric repeat-binding factor 1 | | Authors: | Chen, Y, Yang, Y, van Overbeek, M, Donigian, J.R, Baciu, P, de Lange, T, Lei, M. | | Deposit date: | 2007-12-20 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins.
Science, 319, 2008
|
|
5ZYA
 
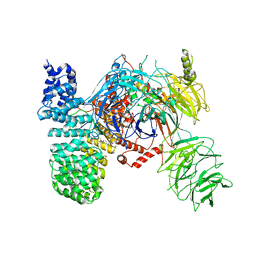 | | SF3b spliceosomal complex bound to E7107 | | Descriptor: | PHD finger-like domain-containing protein 5A, POTASSIUM ION, Splicing factor 3B subunit 1, ... | | Authors: | Finci, L.I, Larsen, N.A. | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | The cryo-EM structure of the SF3b spliceosome complex bound to a splicing modulator reveals a pre-mRNA substrate competitive mechanism of action
Genes Dev., 32, 2018
|
|
8RBX
 
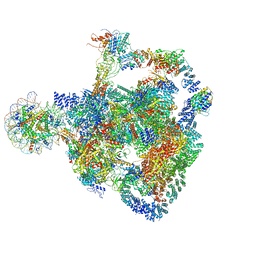 | | Structure of Integrator-PP2A bound to a paused RNA polymerase II-DSIF-NELF-nucleosome complex | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Fianu, I, Ochmann, M, Walshe, J.L, Cramer, P. | | Deposit date: | 2023-12-05 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of Integrator-dependent RNA polymerase II termination.
Nature, 629, 2024
|
|
8RJB
 
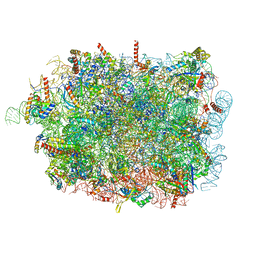 | |
8QX9
 
 | | TDP-43 amyloid fibrils: Morphology-1a | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
8RZV
 
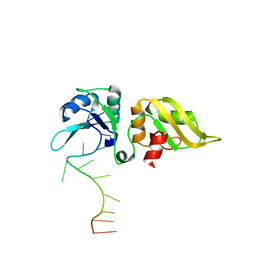 | |
8QOI
 
 | | Structure of the human 80S ribosome at 1.9 A resolution - the molecular role of chemical modifications and ions in RNA | | Descriptor: | 18S rRNA (1740-MER), 28S rRNA (3773-MER), 40S ribosomal protein S10, ... | | Authors: | Holvec, S, Barchet, C, Frechin, L, Hazemann, I, von Loeffelholz, O, Klaholz, B.P. | | Deposit date: | 2023-09-29 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | The structure of the human 80S ribosome at 1.9 angstrom resolution reveals the molecular role of chemical modifications and ions in RNA.
Nat.Struct.Mol.Biol., 2024
|
|
8T4S
 
 | |
8SHE
 
 | | CCT-G beta 5 complex closed state 8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-13 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHD
 
 | | CCT G beta 5 complex closed state 10 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-13 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHP
 
 | | CCT G beta 5 complex closed state 13 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SGL
 
 | | CCT G beta 5 complex closed state 15 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-12 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHA
 
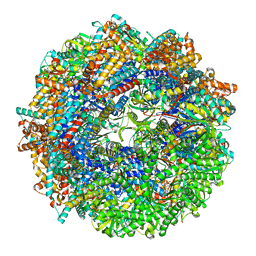 | | CCT-G beta 5 complex closed state 4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-13 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHO
 
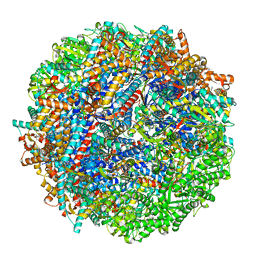 | | CCT G beta 5 complex close state 11 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SFE
 
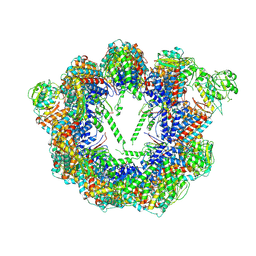 | | Open state CCT-G beta 5 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8QP9
 
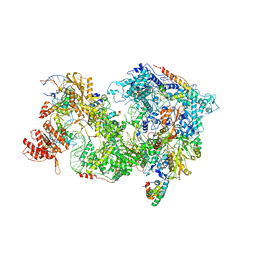 | | Cryo-EM Structure of Pre-B+AMPPNP Complex (core part) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, INOSITOL HEXAKISPHOSPHATE, Pre-mRNA-processing factor 6, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-09-30 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8R08
 
 | | Cryo-EM structure of the cross-exon pre-B+AMPPNP complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, NHP2-like protein 1, N-terminally processed, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8QPE
 
 | | Cryo-EM Structure of Pre-B-like Complex (core part) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, INOSITOL HEXAKISPHOSPHATE, Microfibrillar-associated protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-01 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
