5LMA
 
 | |
7A12
 
 | |
7A2A
 
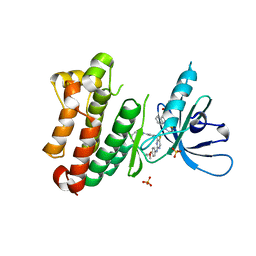 | | Crystal Structure of EGFR-T790M/V948R in Complex with Spebrutinib and EAI001 | | Descriptor: | (2R)-2-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)-2-phenyl-N-(1,3-thiazol-2-yl)acetamide, CHLORIDE ION, Epidermal growth factor receptor, ... | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2020-08-17 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Complex Crystal Structures of EGFR with Third-Generation Kinase Inhibitors and Simultaneously Bound Allosteric Ligands.
Acs Med.Chem.Lett., 11, 2020
|
|
7A3M
 
 | |
7A40
 
 | | Nucleotide-free OSM-3 kinesin motor domain | | Descriptor: | GLYCEROL, Osmotic avoidance abnormal protein 3, SULFATE ION | | Authors: | Varela, F.P, Menetrey, J, Gigant, B. | | Deposit date: | 2020-08-19 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Structural snapshots of the kinesin-2 OSM-3 along its nucleotide cycle: implications for the ATP hydrolysis mechanism.
Febs Open Bio, 11, 2021
|
|
5LHE
 
 | |
6ZYV
 
 | |
6ZZP
 
 | | Crystal structure of (R)-3-hydroxybutyrate dehydrogenase from Psychrobacter arcticus complexed with NAD+ and 3-oxovalerate | | Descriptor: | 3-oxidanylidenepentanoic acid, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative beta-hydroxybutyrate dehydrogenase | | Authors: | Machado, T.F.G, da Silva, R.G, Gloster, T.M, McMahon, S.A, Oehler, V. | | Deposit date: | 2020-08-04 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Dissecting the Mechanism of ( R )-3-Hydroxybutyrate Dehydrogenase by Kinetic Isotope Effects, Protein Crystallography, and Computational Chemistry.
Acs Catalysis, 10, 2020
|
|
7AE2
 
 | | Crystal structure of HEPN(H107A-Y109F) toxin in complex with MNT antitoxin | | Descriptor: | HEPN toxin, MNT ANTITOXIN | | Authors: | Tamulaitiene, G, Sasnauskas, G, Songailiene, I, Juozapaitis, J, Siksnys, V. | | Deposit date: | 2020-09-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HEPN-MNT Toxin-Antitoxin System: The HEPN Ribonuclease Is Neutralized by OligoAMPylation.
Mol.Cell, 80, 2020
|
|
7AE6
 
 | | Crystal structure of di-AMPylated HEPN(R102A) toxin | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, HEPN toxin | | Authors: | Tamulaitiene, G, Sasnauskas, G, Songailiene, I, Juozapaitis, J, Siksnys, V. | | Deposit date: | 2020-09-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | HEPN-MNT Toxin-Antitoxin System: The HEPN Ribonuclease Is Neutralized by OligoAMPylation.
Mol.Cell, 80, 2020
|
|
7A78
 
 | | Crystal structure of RXR beta LBD in complexes with palmitic acid and GRIP-1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Nuclear receptor coactivator 2, ... | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Comprehensive Set of Tertiary Complex Structures and Palmitic Acid Binding Provide Molecular Insights into Ligand Design for RXR Isoforms.
Int J Mol Sci, 21, 2020
|
|
6SI3
 
 | | p53 cancer mutant Y220S in complex with small-molecule stabilizer PK9301 | | Descriptor: | 1,2-ETHANEDIOL, 1-[7-bromanyl-9-[2,2,2-tris(fluoranyl)ethyl]carbazol-3-yl]-~{N}-methyl-methanamine, Cellular tumor antigen p53, ... | | Authors: | Joerger, A.C, Bauer, M.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-08 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Targeting Cavity-Creating p53 Cancer Mutations with Small-Molecule Stabilizers: the Y220X Paradigm.
Acs Chem.Biol., 15, 2020
|
|
5LKE
 
 | |
5LQI
 
 | |
6SIF
 
 | |
6ZT1
 
 | |
6SJ4
 
 | | Amidohydrolase, AHS with substrate analog | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-hydroxyphenyl)carbonyloxybenzoic acid, Amidohydrolase, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7AAS
 
 | | Crystal structure of nitrosoglutathione reductase (GSNOR) from Chlamydomonas reinhardtii | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, S-(hydroxymethyl)glutathione dehydrogenase, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Lemaire, S.D. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional insights into nitrosoglutathione reductase from Chlamydomonas reinhardtii.
Redox Biol, 38, 2020
|
|
6SEX
 
 | | X-ray structure of the gold/lysozyme adduct formed upon 21h exposure of protein crystals to compound 1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Ferraro, G, Giorgio, A, Merlino, A. | | Deposit date: | 2019-07-30 | | Release date: | 2019-09-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Protein-mediated disproportionation of Au(i): insights from the structures of adducts of Au(iii) compounds bearing N,N-pyridylbenzimidazole derivatives with lysozyme.
Dalton Trans, 48, 2019
|
|
6SFH
 
 | | CRYSTAL STRUCTURE OF DHQ1 FROM Staphylococcus aureus COVALENTLY MODIFIED BY LIGAND 7 | | Descriptor: | (1~{S},3~{S},4~{S},5~{R})-3-(aminomethyl)-3,4,5-tris(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase, LITHIUM ION, ... | | Authors: | Sanz-Gaitero, M, Lence, E, Maneiro, M, Thompson, R, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Self-Immolation of a Bacterial Dehydratase Enzyme by its Epoxide Product.
Chemistry, 26, 2020
|
|
5LSB
 
 | | Crystal structure of yeast Hsh49p in complex with Cus1p binding domain. | | Descriptor: | Cold sensitive U2 snRNA suppressor 1, Protein HSH49 | | Authors: | van Roon, A.M, Obayashi, E, Sposito, B, Oubridge, C, Nagai, K. | | Deposit date: | 2016-08-24 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of U2 snRNP SF3b components: Hsh49p in complex with Cus1p-binding domain.
RNA, 23, 2017
|
|
7AED
 
 | | VirB8 domain of PrgL from Enterococcus faecalis pCF10 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PrgL | | Authors: | Jaeger, F, Berntsson, R.P.A. | | Deposit date: | 2020-09-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Structure of the enterococcal T4SS protein PrgL reveals unique dimerization interface in the VirB8 protein family.
Structure, 30, 2022
|
|
5LSP
 
 | | 107_A07 Fab in complex with fragment of the Met receptor | | Descriptor: | 107_A07 Fab heavy chain, 107_A07 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | DiCara, D, Chirgadze, D.Y, Pope, A, Karatt-Vellatt, A, Winter, A, van den Heuvel, J, Gherardi, E, McCafferty, J. | | Deposit date: | 2016-09-05 | | Release date: | 2017-09-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Characterization and structural determination of a new anti-MET function-blocking antibody with binding epitope distinct from the ligand binding domain.
Sci Rep, 7, 2017
|
|
6SKV
 
 | |
6ZZQ
 
 | | Crystal structure of (R)-3-hydroxybutyrate dehydrogenase from Acinetobacter baumannii complexed with NAD+ and acetoacetate | | Descriptor: | 3-hydroxybutyrate dehydrogenase, ACETOACETIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Machado, T.F.G, da Silva, R.G, Gloster, T.M, McMahon, S.A, Oehler, V. | | Deposit date: | 2020-08-05 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Dissecting the Mechanism of ( R )-3-Hydroxybutyrate Dehydrogenase by Kinetic Isotope Effects, Protein Crystallography, and Computational Chemistry.
Acs Catalysis, 10, 2020
|
|
