4K22
 
 | | Structure of the C-terminal truncated form of E.Coli C5-hydroxylase UBII involved in ubiquinone (Q8) biosynthesis | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Pecqueur, L, Lombard, M, Golinelli-pimpaneau, B, Fontecave, M. | | Deposit date: | 2013-04-07 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ubiI, a New Gene in Escherichia coli Coenzyme Q Biosynthesis, Is Involved in Aerobic C5-hydroxylation.
J.Biol.Chem., 288, 2013
|
|
5IFS
 
 | | Quantitative interaction mapping reveals an extended ubiquitin regulatory domain in ASPL that disrupts functional p97 hexamers and induces cell death | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Roske, Y, Arumughan, A, Heinemann, U, Wanker, E. | | Deposit date: | 2016-02-26 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Quantitative interaction mapping reveals an extended UBX domain in ASPL that disrupts functional p97 hexamers.
Nat Commun, 7, 2016
|
|
1MVN
 
 | | PPC decarboxylase mutant C175S complexed with pantothenoylaminoethenethiol | | Descriptor: | 2,4-DIHYDROXY-N-[2-(2-MERCAPTO-VINYLCARBAMOYL)-ETHYL]-3,3-DIMETHYL-BUTYRAMIDE, FLAVIN MONONUCLEOTIDE, PPC decarboxylase AtHAL3a | | Authors: | Steinbacher, S, Hernandez-Acosta, P, Bieseler, B, Blaesse, M, Huber, R, Culianez-Macia, F.A, Kupke, T. | | Deposit date: | 2002-09-26 | | Release date: | 2003-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal Structure of the Plant PPC Decarboxylase AtHAL3a Complexed with an Ene-thiol Reaction Intermediate
J.Mol.Biol., 327, 2003
|
|
6PCA
 
 | | Crystal structure of beta-ketoadipyl-CoA thiolase | | Descriptor: | ACETATE ION, Beta-ketoadipyl-CoA thiolase, CHLORIDE ION, ... | | Authors: | Sukritee, B, Panjikar, S. | | Deposit date: | 2019-06-17 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for differentiation between two classes of thiolase: Degradative vs biosynthetic thiolase.
J Struct Biol X, 4, 2020
|
|
4QGI
 
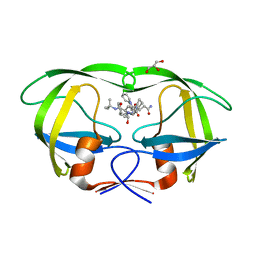 | | X-ray crystal structure of HIV-1 protease variant G48T/L89M in complex with Saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, GLYCEROL, Protease | | Authors: | Mahon, B.P, McKenna, R, Goldfarb, N. | | Deposit date: | 2014-05-22 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Defective Hydrophobic Sliding Mechanism and Active Site Expansion in HIV-1 Protease Drug Resistant Variant Gly48Thr/Leu89Met: Mechanisms for the Loss of Saquinavir Binding Potency.
Biochemistry, 54, 2015
|
|
4QEV
 
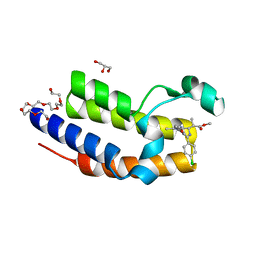 | | Crystal structure of BRD2(BD2) mutant with ligand ME bound (METHYL (2R)- 2-[(4S)-6-(4-CHLOROPHENYL)-8-METHOXY-1-METHYL-4H-[1,2,4]TRIAZOLO[4,3-A][1, 4]BENZODIAZEPIN-4-YL]PROPANOATE) | | Descriptor: | Bromodomain-containing protein 2, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Tallant, C, Baud, M, Lin-Shiao, E, Chirgadze, D.Y, Ciulli, A. | | Deposit date: | 2014-05-19 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemical biology. A bump-and-hole approach to engineer controlled selectivity of BET bromodomain chemical probes.
Science, 346, 2014
|
|
6PCB
 
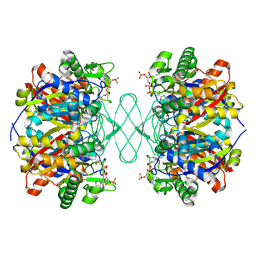 | |
5QJ1
 
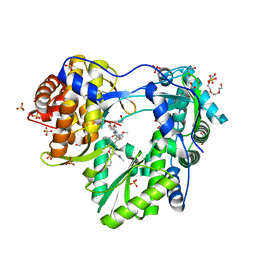 | |
6P90
 
 | | Crystal structure of PaDHDPS2-H56Q mutant | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, CHLORIDE ION, GLYCEROL | | Authors: | Impey, R.E, Panjikar, S, Hall, C.J, Bock, L.J, Sutton, J.M, Perugini, M.A, Soares da Costa, T.P. | | Deposit date: | 2019-06-08 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of two dihydrodipicolinate synthase isoforms from Pseudomonas aeruginosa that differ in allosteric regulation.
Febs J., 287, 2020
|
|
6PCC
 
 | |
6PCD
 
 | |
8KCM
 
 | | MmCPDII-DNA complex containing low-dosage, light induced repaired DNA. | | Descriptor: | Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
8K2K
 
 | | Crystal structure of Group 3 Oligosaccharide/Monosaccharide-releasing beta-N-acetylgalactosaminidase NgaDssm in complex with GalNAc-thiazoline | | Descriptor: | (3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methyl-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sumida, T, Fushinobu, S. | | Deposit date: | 2023-07-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Genetic and functional diversity of beta-N-acetylgalactosamine-targeting glycosidases expanded by deep-sea metagenome analysis.
Nat Commun, 15, 2024
|
|
5QJ0
 
 | |
4RLQ
 
 | | Crystal structure of a benzoate coenzyme A ligase with o-Toluic acid | | Descriptor: | 2-methylbenzoic acid, Benzoate-coenzyme A ligase, GLYCEROL | | Authors: | Strom, S, Nosrati, M, Thornburg, C, Walker, K.D, Geiger, J.H. | | Deposit date: | 2014-10-17 | | Release date: | 2015-09-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Kinetically and Crystallographically Guided Mutations of a Benzoate CoA Ligase (BadA) Elucidate Mechanism and Expand Substrate Permissivity.
Biochemistry, 54, 2015
|
|
4RLF
 
 | | Crystal structure of a benzoate coenzyme A ligase with p-Toluic acid and o-Toluic acid | | Descriptor: | 2-methylbenzoic acid, 4-METHYLBENZOIC ACID, Benzoate-coenzyme A ligase, ... | | Authors: | Strom, S, Nosrati, M, Thornburg, C, Walker, K, Geiger, J.H. | | Deposit date: | 2014-10-16 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Kinetically and Crystallographically Guided Mutations of a Benzoate CoA Ligase (BadA) Elucidate Mechanism and Expand Substrate Permissivity.
Biochemistry, 54, 2015
|
|
4RMN
 
 | | Crystal structure of a benzoate coenzyme A ligase with 2-Thiophene Carboxylic acid | | Descriptor: | Benzoate-coenzyme A ligase, GLYCEROL, THIOPHENE-2-CARBOXYLIC ACID | | Authors: | Strom, S, Nosrati, M, Thornburg, C, Walker, K.D, Geiger, J.H. | | Deposit date: | 2014-10-21 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Kinetically and Crystallographically Guided Mutations of a Benzoate CoA Ligase (BadA) Elucidate Mechanism and Expand Substrate Permissivity.
Biochemistry, 54, 2015
|
|
4OIM
 
 | | Crystal structure of Mycobacterium tuberculosis InhA in complex with inhibitor PT119 in 2.4 M acetate | | Descriptor: | 2-(2-CYANOPHENOXY)-5-HEXYLPHENOL, ACETATE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-01-20 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Time-Dependent Diaryl Ether Inhibitors of InhA: Structure-Activity Relationship Studies of Enzyme Inhibition, Antibacterial Activity, and in vivo Efficacy.
Chemmedchem, 9, 2014
|
|
4HZN
 
 | | The Structure of the Bifunctional Acetyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double Hot Dog Fold | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional Methylmalonyl-CoA:ACP Acyltransferase/Decarboxylase, GLYCEROL, ... | | Authors: | Lohman, J.R, Bingman, C.A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-15 | | Release date: | 2013-01-30 | | Last modified: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the Bifunctional Acyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double-Hot-Dog Fold.
Biochemistry, 52, 2013
|
|
1ELT
 
 | |
6B8X
 
 | | Multiconformer model of apo WT PTP1B with glycerol at 278 K | | Descriptor: | GLYCEROL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2017-10-09 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
4Q9F
 
 | | Crystal structure of type 1 ribosome inactivating protein from Momordica balsamina in complex with guanosine mono phosphate at 1.75 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Kushwaha, G.S, Pandey, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-05-01 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of type 1 ribosome inactivating protein from Momordica balsamina in complex with guanosine mono phosphate at 1.75 Angstrom resolution
To be Published
|
|
1S5S
 
 | | Porcine trypsin complexed with guanidine-3-propanol inhibitor | | Descriptor: | CALCIUM ION, GLYCEROL, GUANIDINE-3-PROPANOL, ... | | Authors: | Transue, T.R, Krahn, J.M, Gabel, S.A, DeRose, E.F, London, R.E. | | Deposit date: | 2004-01-21 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray and NMR characterization of covalent complexes of trypsin, borate, and alcohols.
Biochemistry, 43, 2004
|
|
6CU6
 
 | |
4RBP
 
 | | Crystal structure of HIV neutralizing antibody 2G12 in complex with a bacterial oligosaccharide analog of mammalian oligomanose | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, GLYCEROL, ... | | Authors: | Stanfield, R.L, Wilson, I.A, De Castro, C, Marzaioli, A.M, Pantophlet, R. | | Deposit date: | 2014-09-12 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the HIV neutralizing antibody 2G12 in complex with a bacterial oligosaccharide analog of mammalian oligomannose.
Glycobiology, 25, 2015
|
|
