5UP4
 
 | |
2W0T
 
 | |
5UIY
 
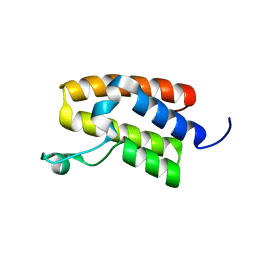 | | Structure of Bromodomain from human BAZ1A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bromodomain adjacent to zinc finger domain protein 1A | | Authors: | Oppikofer, M, Sudhamsu, J. | | Deposit date: | 2017-01-16 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.687 Å) | | Cite: | Non-canonical reader modules of BAZ1A promote recovery from DNA damage.
Nat Commun, 8, 2017
|
|
2X0U
 
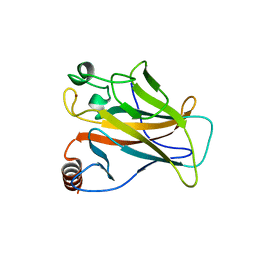 | | STRUCTURE OF THE P53 CORE DOMAIN MUTANT Y220C BOUND TO A 2-amino substituted benzothiazole scaffold | | Descriptor: | 6,7-DIHYDRO[1,4]DIOXINO[2,3-F][1,3]BENZOTHIAZOL-2-AMINE, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Joerger, A.C, Kaar, J.L, Basse, N, Fersht, A.R. | | Deposit date: | 2009-12-17 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Toward the Rational Design of P53-Stabilizing Drugs: Probing the Surface of the Oncogenic Y220C Mutant.
Chem.Biol., 17, 2010
|
|
5SUY
 
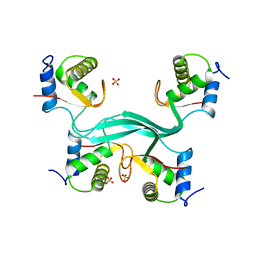 | |
2VUK
 
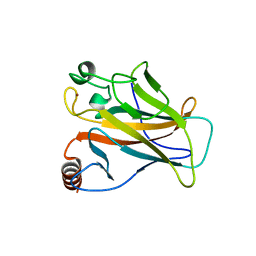 | |
2X0V
 
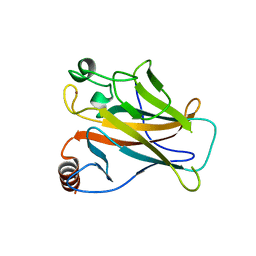 | | STRUCTURE OF THE P53 CORE DOMAIN MUTANT Y220C BOUND TO 4-(trifluoromethyl)benzene-1,2-diamine | | Descriptor: | 4-(TRIFLUOROMETHYL)BENZENE-1,2-DIAMINE, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Basse, N, Kaar, J.L, Joerger, A.C, Fersht, A.R. | | Deposit date: | 2009-12-17 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward the Rational Design of P53-Stabilizing Drugs: Probing the Surface of the Oncogenic Y220C Mutant.
Chem.Biol., 17, 2010
|
|
5TOI
 
 | |
5UQV
 
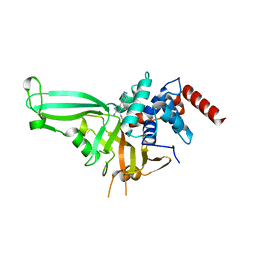 | |
5UJ2
 
 | | Crystal structure of HCV NS5B genotype 2A JFH-1 isolate with S15G E86Q E87Q C223H V321I mutations and Delta8 neta hairpoin loop deletion in complex with GS-639476 (diphsohate version of GS-9813), Mn2+ and symmetrical primer template 5'-AUAAAUUU | | Descriptor: | (1S)-1-(4-aminoimidazo[2,1-f][1,2,4]triazin-7-yl)-1,4-anhydro-2-deoxy-2-fluoro-5-O-[(S)-hydroxy(phosphonooxy)phosphoryl]-2-methyl-D-ribitol, CHLORIDE ION, Genome polyprotein, ... | | Authors: | Edwards, T.E, Fox III, D, Appleby, T.C, Murakami, E, Rey, A, McGrath, M.E. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of a 2'-fluoro-2'-C-methyl C-nucleotide HCV polymerase inhibitor and a phosphoramidate prodrug with favorable properties.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5UQX
 
 | |
5THL
 
 | |
5THH
 
 | |
5VD6
 
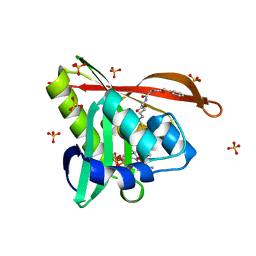 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with bisubstrate analog 6 | | Descriptor: | (3R,5S,9R,23S)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-10,14-dioxo-23-({[(phenylacetyl)amino]acetyl}amino)-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphatetracosan-24-oic acid 3,5-dioxide (non-preferred name), SULFATE ION, acetyltransferase PA4794 | | Authors: | Majorek, K.A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-04-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Generating enzyme and radical-mediated bisubstrates as tools for investigating Gcn5-related N-acetyltransferases.
FEBS Lett., 591, 2017
|
|
8ERF
 
 | |
6ERF
 
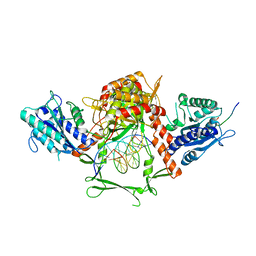 | | Complex of APLF factor and Ku heterodimer bound to DNA | | Descriptor: | Aprataxin and PNK-like factor, DNA (34-MER), DNA (5'-D(*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*TP*TP*GP*GP*GP*CP*GP*CP*G)-3'), ... | | Authors: | Nemoz, C, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2017-10-18 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining.
Nat.Struct.Mol.Biol., 25, 2018
|
|
5ERF
 
 | |
6ERG
 
 | | Complex of XLF and heterodimer Ku bound to DNA | | Descriptor: | DNA (21-MER), DNA (34-MER), Non-homologous end-joining factor 1, ... | | Authors: | Nemoz, C, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2017-10-18 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4ERF
 
 | | crystal structure of MDM2 (17-111) in complex with compound 29 (AM-8553) | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, {(3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-1-[(2S,3S)-2-hydroxypentan-3-yl]-3-methyl-2-oxopiperidin-3-yl}acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel inhibitors of the MDM2-p53 interaction.
J.Med.Chem., 55, 2012
|
|
3ERF
 
 | | Crystal structure of Gtt2 from Saccharomyces cerevisiae | | Descriptor: | Glutathione S-transferase 2 | | Authors: | Ma, X.X, Jiang, Y.L, He, Y.X, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2008-10-02 | | Release date: | 2009-10-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structures of yeast glutathione-S-transferase Gtt2 reveal a new catalytic type of GST family.
Embo Rep., 10, 2009
|
|
4ERE
 
 | | crystal structure of MDM2 (17-111) in complex with compound 23 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(3R,5R,6S)-1-[(2S)-1-tert-butoxy-1-oxobutan-2-yl]-5-(3-chlorophenyl)-6-(4-chlorophenyl)-2-oxopiperidin-3-yl]acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of novel inhibitors of the MDM2-p53 interaction.
J.Med.Chem., 55, 2012
|
|
3ERG
 
 | | Crystal structure of Gtt2 from Saccharomyces cerevisiae in complex with glutathione sulfnate | | Descriptor: | GLUTATHIONE SULFONIC ACID, Glutathione S-transferase 2 | | Authors: | Ma, X.X, Jiang, Y.L, He, Y.X, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2008-10-02 | | Release date: | 2009-10-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of yeast glutathione-S-transferase Gtt2 reveal a new catalytic type of GST family.
Embo Rep., 10, 2009
|
|
6TZH
 
 | | ADC-7 in complex with boronic acid transition state inhibitor S06015 | | Descriptor: | Beta-lactamase, GLYCINE, PHOSPHATE ION, ... | | Authors: | Fish, E.R, Powers, R.A, Wallar, B.J. | | Deposit date: | 2019-08-12 | | Release date: | 2020-06-24 | | Last modified: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | 1,2,3-Triazolylmethaneboronate: A Structure Activity Relationship Study of a Class of beta-Lactamase Inhibitors againstAcinetobacter baumanniiCephalosporinase.
Acs Infect Dis., 6, 2020
|
|
5K2G
 
 | |
5K2H
 
 | |
