6FH5
 
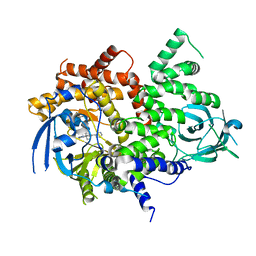 | | PI3Kg IN COMPLEX WITH Compound 7 | | Descriptor: | 3-methyl-1-(oxan-4-yl)-8-pyridin-3-yl-imidazo[4,5-c]quinolin-2-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Petersen, J, Barlaam, B. | | Deposit date: | 2018-01-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery and Optimisation of a Novel Series of 3-Cinnoline Carboxamides as Orally Bioavailable, Highly Potent and Selective Ataxia Telangiectasia Mutated (ATM) inhibitors
To Be Published
|
|
7Q0F
 
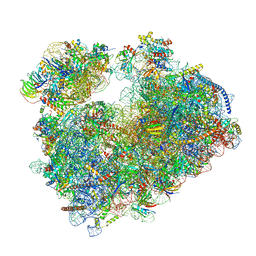 | | Structure of Candida albicans 80S ribosome in complex with phyllanthoside | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Zgadzay, Y, Kolosova, O, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-14 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
7PZY
 
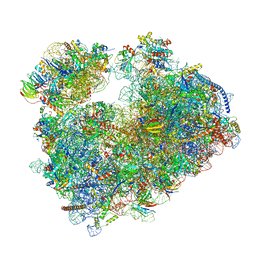 | | Structure of the vacant Candida albicans 80S ribosome | | Descriptor: | 1,4-DIAMINOBUTANE, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Zgadzay, Y, Kolosova, O, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-13 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
7Q0P
 
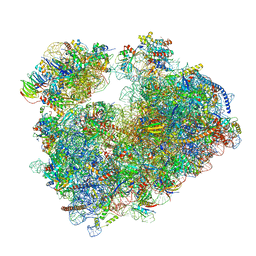 | | Structure of the Candida albicans 80S ribosome in complex with anisomycin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Kolosova, O, Zgadzay, Y, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-15 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
7Q08
 
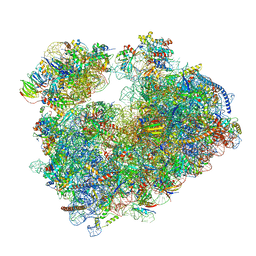 | | Structure of Candida albicans 80S ribosome in complex with cycloheximide | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Zgadzay, Y, Kolosova, O, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-14 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
4BY2
 
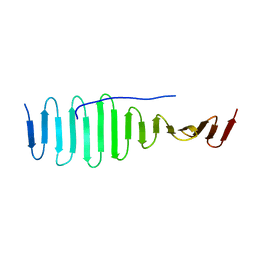 | | SAS-4 (dCPAP) TCP domain in complex with a Proline Rich Motif of Ana2 (dSTIL) of Drosophila Melanogaster | | Descriptor: | 1,2-ETHANEDIOL, ANASTRAL SPINDLE 2, SAS 4 | | Authors: | Cottee, M.A, Muschalik, N, Wong, Y.L, Johnson, C.M, Johnson, S, Andreeva, A, Oegema, K, Lea, S.M, Raff, J.W, van Breugel, M. | | Deposit date: | 2013-07-17 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal structures of the CPAP/STIL complex reveal its role in centriole assembly and human microcephaly.
Elife, 2, 2013
|
|
7R63
 
 | |
4BUZ
 
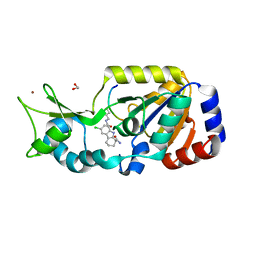 | | SIR2 COMPLEX STRUCTURE MIXTURE OF EX-527 INHIBITOR AND REACTION PRODUCTS OR OF REACTION SUBSTRATES P53 PEPTIDE AND NAD | | Descriptor: | (1S)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1- carboxamide, 1,2-ETHANEDIOL, 2'-O-ACETYL ADENOSINE-5-DIPHOSPHORIBOSE, ... | | Authors: | Weyand, M, Lakshminarasimhan, M, Gertz, M, Steegborn, C. | | Deposit date: | 2013-06-24 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8DP5
 
 | | Structure of the PEAK3/14-3-3 complex | | Descriptor: | 14-3-3 protein beta/alpha, 14-3-3 protein epsilon, Protein PEAK3, ... | | Authors: | Torosyan, H, Paul, M, Jura, N, Verba, K.A. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into regulation of the PEAK3 pseudokinase scaffold by 14-3-3.
Nat Commun, 14, 2023
|
|
8DS6
 
 | |
7R55
 
 | | B-trefoil lectin from Salpingoeca rosetta in complex with Gb3 | | Descriptor: | 1,2-ETHANEDIOL, Sarol-1, alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Notova, S, Varrot, A. | | Deposit date: | 2022-02-10 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The choanoflagellate pore-forming lectin SaroL-1 punches holes in cancer cells by targeting the tumor-related glycosphingolipid Gb3.
Commun Biol, 5, 2022
|
|
4DG7
 
 | |
7RRW
 
 | | Monomeric CRM197 expressed in E. coli | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Diphtheria toxin | | Authors: | Gallagher, D.T, Lees, A. | | Deposit date: | 2021-08-10 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Monomeric crystal structure of the vaccine carrier protein CRM 197 and implications for vaccine development.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8EW6
 
 | | Anti-human CD8 VHH complex with CD8 alpha | | Descriptor: | Anti-CD8 alpha VHH, T-cell surface glycoprotein CD8 alpha chain | | Authors: | Kiefer, J.R, Williams, S, Davies, C.W, Koerber, J.T, Sriraman, S.K, Yin, Y.P. | | Deposit date: | 2022-10-21 | | Release date: | 2022-11-23 | | Last modified: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Development of an 18 F-labeled anti-human CD8 VHH for same-day immunoPET imaging.
Eur J Nucl Med Mol Imaging, 50, 2023
|
|
1D2H
 
 | | CRYSTAL STRUCTURE OF R175K MUTANT GLYCINE N-METHYLTRANSFERASE COMPLEXED WITH S-ADENOSYLHOMOCYSTEINE | | Descriptor: | GLYCINE N-METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Huang, Y, Komoto, J, Takusagawa, F, Konishi, K, Takata, Y. | | Deposit date: | 1999-10-11 | | Release date: | 1999-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanisms for auto-inhibition and forced product release in glycine N-methyltransferase: crystal structures of wild-type, mutant R175K and S-adenosylhomocysteine-bound R175K enzymes.
J.Mol.Biol., 298, 2000
|
|
1AA5
 
 | | VANCOMYCIN | | Descriptor: | ACETIC ACID, CHLORIDE ION, VANCOMYCIN, ... | | Authors: | Loll, P.J, Bevivino, A.E, Korty, B.D, Axelsen, P.H. | | Deposit date: | 1997-01-23 | | Release date: | 1997-08-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Simultaneous Recognition of a Carboxylate-Containing Ligand and an Intramolecular Surrogate Ligand in the Crystal Structure of an Asymmetric Vancomycin Dimer.
J.Am.Chem.Soc., 119, 1997
|
|
6ES7
 
 | |
6EFH
 
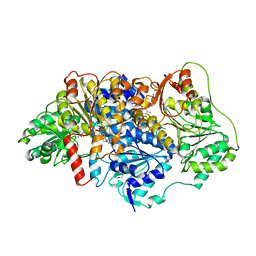 | | Pyruvate decarboxylase from Kluyveromyces lactis soaked with pyruvamide | | Descriptor: | (1S,2S)-1-amino-1,2-dihydroxypropan-1-olate, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Kutter, S, Konig, S. | | Deposit date: | 2018-08-16 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The crystal structures of pyruvate decarboxylase from Kluyveromyces lactis in the absence of ligands and in the presence of the substrate surrogate pyruvamide
To be Published
|
|
1BG9
 
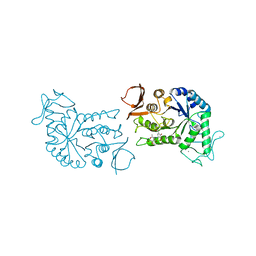 | | BARLEY ALPHA-AMYLASE WITH SUBSTRATE ANALOGUE ACARBOSE | | Descriptor: | 1,4-ALPHA-D-GLUCAN GLUCANOHYDROLASE, 4,6-dideoxy-4-{[(1S,4S,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Kadziola, A, Haser, R. | | Deposit date: | 1998-06-05 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular structure of a barley alpha-amylase-inhibitor complex: implications for starch binding and catalysis.
J.Mol.Biol., 278, 1998
|
|
1DYQ
 
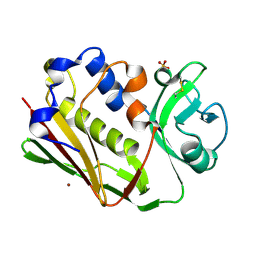 | | STAPHYLOCOCCAL ENTEROTOXIN A MUTANT VACCINE | | Descriptor: | Enterotoxin type A, SULFATE ION, ZINC ION | | Authors: | Krupka, H.I, Segelke, B.W, Rupp, B. | | Deposit date: | 2000-02-05 | | Release date: | 2001-03-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Abrogated Binding between Staphylococcal Enterotoxin a Superantigen Vaccine and Mhc-II?
Protein Sci., 11, 2002
|
|
4GEL
 
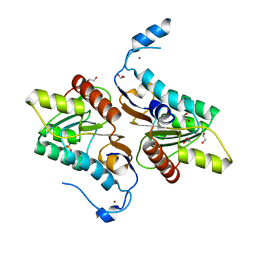 | | Crystal structure of Zucchini | | Descriptor: | 1,2-ETHANEDIOL, Mitochondrial cardiolipin hydrolase, PHOSPHATE ION, ... | | Authors: | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
4G6W
 
 | | Human Thymidylate Synthase M190K with bound 4-Bromobenzene-1,2,3-triol | | Descriptor: | 4-bromobenzene-1,2,3-triol, SULFATE ION, Thymidylate synthase | | Authors: | Celeste, L.R, Lebioda, L. | | Deposit date: | 2012-07-19 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Oxidation of Cysteine 195 of Huyman Thymidylate Synthase by Purpurogallin
To be Published, 2012
|
|
1D2C
 
 | | METHYLTRANSFERASE | | Descriptor: | PROTEIN (GLYCINE N-METHYLTRANSFERASE) | | Authors: | Huang, Y, Takusagawa, F. | | Deposit date: | 1999-09-23 | | Release date: | 1999-10-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanisms for auto-inhibition and forced product release in glycine N-methyltransferase: crystal structures of wild-type, mutant R175K and S-adenosylhomocysteine-bound R175K enzymes.
J.Mol.Biol., 298, 2000
|
|
6M26
 
 | | Sirohydrochlorin nickelochelatase CfbA in P212121 space group | | Descriptor: | Sirohydrochlorin cobaltochelatase | | Authors: | Fujishiro, T. | | Deposit date: | 2020-02-26 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The nickel-sirohydrochlorin formation mechanism of the ancestral class II chelatase CfbA in coenzyme F430 biosynthesis.
Chem Sci, 12, 2021
|
|
6M29
 
 | |
