1TK5
 
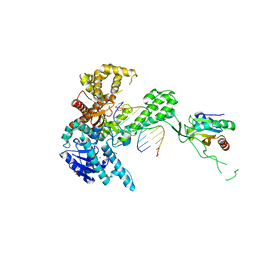 | | T7 DNA polymerase binary complex with 8 oxo guanosine in the templating strand | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(*CP*CP*CP*(8OG)P*CP*TP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*TP*CP*G)-3', 5'-D(*CP*GP*AP*AP*A*GP*CP*CP*AP*GP*TP*GP*CP*CP*AP*(DDG)P*TP*GP*CP*AP*A)-3', ... | | Authors: | Brieba, L.G, Eichman, B.F, Kokoska, R.J, Doublie, S, Kunkel, T.A, Ellenberger, T. | | Deposit date: | 2004-06-08 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase.
Embo J., 23, 2004
|
|
1TK0
 
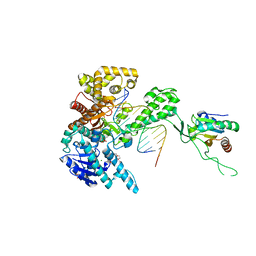 | | T7 DNA polymerase ternary complex with 8 oxo guanosine and ddCTP at the insertion site | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(*CP*CP*CP*(8OG)P*CP*TP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*TP*CP*G)-3', ... | | Authors: | Brieba, L.G, Eichman, B.F, Kokoska, R.J, Doublie, S, Kunkel, T.A, Ellenberger, T. | | Deposit date: | 2004-06-07 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase.
Embo J., 23, 2004
|
|
6XAR
 
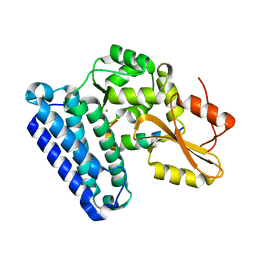 | |
5JBK
 
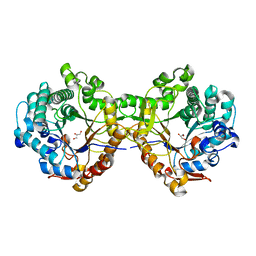 | | Trichoderma harzianum GH1 beta-glucosidase ThBgl1 | | Descriptor: | Beta-glucosidase, GLYCEROL | | Authors: | Florindo, R.N, Mutti, H.S, Polikarpov, I, Nascimento, A.S. | | Deposit date: | 2016-04-13 | | Release date: | 2017-08-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Structural insights into beta-glucosidase transglycosylation based on biochemical, structural and computational analysis of two GH1 enzymes from Trichoderma harzianum.
N Biotechnol, 40, 2018
|
|
1TKD
 
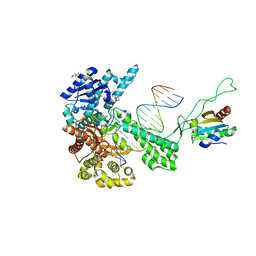 | | T7 DNA polymerase ternary complex with 8 oxo guanosine and dCMP at the elongation site | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*CP*CP*CP*AP*(8OG)P*TP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*TP*CP*G)-3'), ... | | Authors: | Brieba, L.G, Eichman, B.F, Kokoska, R.J, Doublie, S, Kunkel, T.A, Ellenberger, T. | | Deposit date: | 2004-06-08 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase.
Embo J., 23, 2004
|
|
6LUR
 
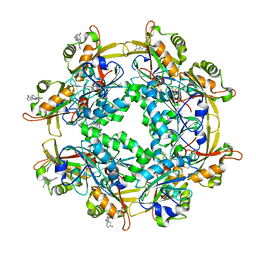 | |
2EIO
 
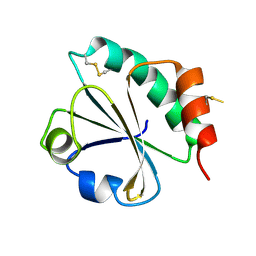 | |
2EIQ
 
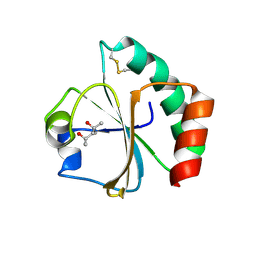 | |
6IBL
 
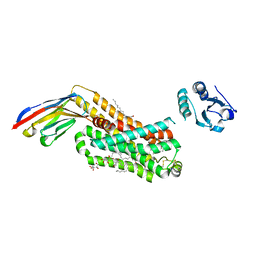 | | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND AGONIST FORMOTEROL AND NANOBODY Nb80 | | Descriptor: | Camelid antibody fragment Nb80, HEGA-10, SODIUM ION, ... | | Authors: | Warne, T, Edwards, P.C, Dore, A.S, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of beta-arrestin coupling to formoterol-bound beta1-adrenoceptor.
Nature, 583, 2020
|
|
6H7L
 
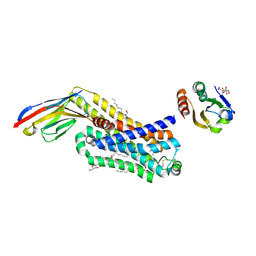 | | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND PARTIAL AGONIST DOBUTAMINE AND NANOBODY Nb6B9 | | Descriptor: | Beta-1 adrenergic receptor, Camelid antibody fragment Nb6B9, DOBUTAMINE, ... | | Authors: | Warne, T, Edwards, P.C, Dore, A.S, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2018-07-31 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for high-affinity agonist binding in GPCRs.
Science, 364, 2019
|
|
6H7O
 
 | | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND WEAK PARTIAL AGONIST CYANOPINDOLOL AND NANOBODY Nb6B9 | | Descriptor: | 4-{[(2S)-3-(tert-butylamino)-2-hydroxypropyl]oxy}-3H-indole-2-carbonitrile, Beta-1 adrenergic receptor, Camelid antibody fragment Nb6B9, ... | | Authors: | Warne, T, Edwards, P.C, Dore, A.S, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2018-07-31 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for high-affinity agonist binding in GPCRs.
Science, 364, 2019
|
|
3AQX
 
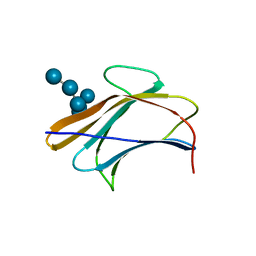 | | Crystal structure of Bombyx mori beta-GRP/GNBP3 N-terminal domain with laminarihexaoses | | Descriptor: | Beta-1,3-glucan-binding protein, GLYCEROL, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Kanagawa, M, Satoh, T, Ikeda, A, Adachi, Y, Ohno, N, Yamaguchi, Y. | | Deposit date: | 2010-11-22 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into recognition of triple-helical beta-glucans by an insect fungal receptor
J.Biol.Chem., 286, 2011
|
|
6H7M
 
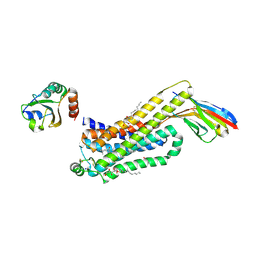 | | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND PARTIAL AGONIST SALBUTAMOL AND NANOBODY Nb6B9 | | Descriptor: | Beta-1 adrenergic receptor, Camelid antibody fragment Nb6B9, HEGA-10, ... | | Authors: | Warne, T, Edwards, P.C, Dore, A.S, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2018-07-31 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Molecular basis for high-affinity agonist binding in GPCRs.
Science, 364, 2019
|
|
6H7N
 
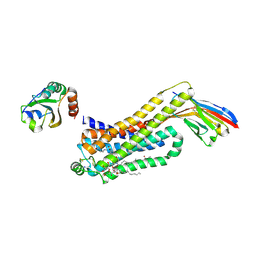 | | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND PARTIAL AGONIST XAMOTEROL AND NANOBODY Nb6B9 | | Descriptor: | Beta-1 adrenergic receptor, Camelid antibody fragment Nb6B9, HEGA-10, ... | | Authors: | Warne, T, Edwards, P.C, Dore, A.S, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2018-07-31 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for high affinity agonist binding in GPCRs
Biorxiv, 2018
|
|
3AQY
 
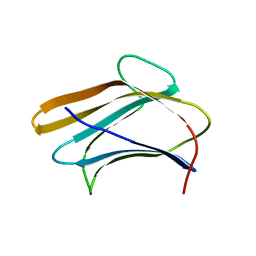 | | Crystal structure of Plodia interpunctella beta-GRP/GNBP3 N-terminal domain | | Descriptor: | Beta-1,3-glucan-binding protein | | Authors: | Kanagawa, M, Satoh, T, Ikeda, A, Adachi, Y, Ohno, N, Yamaguchi, Y. | | Deposit date: | 2010-11-22 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural insights into recognition of triple-helical beta-glucans by an insect fungal receptor
J.Biol.Chem., 286, 2011
|
|
3AQZ
 
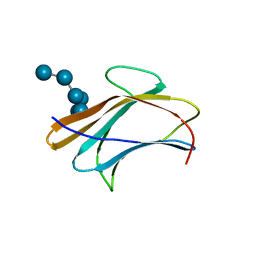 | | Crystal structure of Plodia interpunctella beta-GRP/GNBP3 N-terminal domain with laminarihexaoses | | Descriptor: | Beta-1,3-glucan-binding protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Kanagawa, M, Satoh, T, Ikeda, A, Adachi, Y, Ohno, N, Yamaguchi, Y. | | Deposit date: | 2010-11-22 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into recognition of triple-helical beta-glucans by an insect fungal receptor
J.Biol.Chem., 286, 2011
|
|
5E4W
 
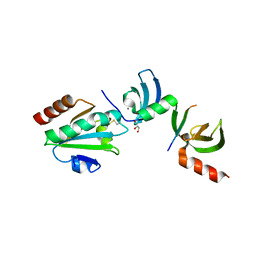 | | Crystal structure of cpSRP43 chromodomains 2 and 3 in complex with the Alb3 tail | | Descriptor: | CALCIUM ION, GLYCEROL, Inner membrane protein ALBINO3, ... | | Authors: | Horn, A, Ahmed, Y.L, Wild, K, Sinning, I. | | Deposit date: | 2015-10-07 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for cpSRP43 chromodomain selectivity and dynamics in Alb3 insertase interaction.
Nat Commun, 6, 2015
|
|
6H7J
 
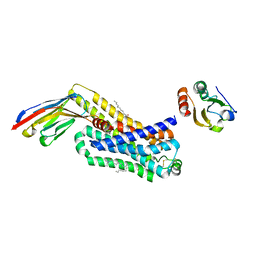 | | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND AGONIST ISOPRENALINE AND NANOBODY Nb80 | | Descriptor: | Beta-1 adrenergic receptor, Camelid antibody fragment Nb80, HEGA-10, ... | | Authors: | Warne, T, Edwards, P.C, Dore, A.S, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2018-07-31 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for high-affinity agonist binding in GPCRs.
Science, 364, 2019
|
|
6TRV
 
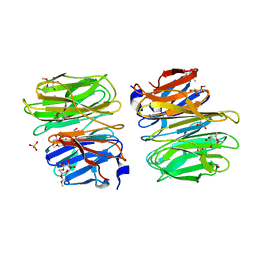 | |
7V8H
 
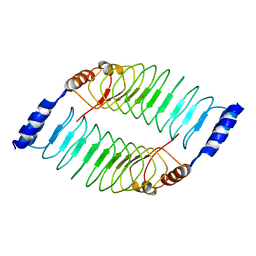 | | Crystal structure of LRR domain from Shigella flexneri IpaH1.4 | | Descriptor: | RING-type E3 ubiquitin transferase | | Authors: | Liu, J, Wang, Y, Pan, L. | | Deposit date: | 2021-08-23 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Mechanistic insights into the subversion of the linear ubiquitin chain assembly complex by the E3 ligase IpaH1.4 of Shigella flexneri.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7JST
 
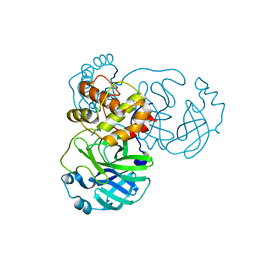 | | Crystal structure of SARS-CoV-2 3CL in apo form | | Descriptor: | 3C-like proteinase, PHOSPHATE ION | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Huang, Y, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-16 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
7ALD
 
 | |
4E8H
 
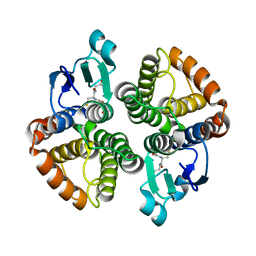 | | Structural of Bombyx mori glutathione transferase BmGSTD1 complex with GTT | | Descriptor: | GLUTATHIONE, Glutathione S-transferase | | Authors: | Tan, X, Ma, X.X, Hu, X.M, Chen, Q.M, Zhao, P, Xia, Q.Y, Zhou, C.Z. | | Deposit date: | 2012-03-20 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural of Bombyx mori glutathione transferase BmGSTD1 complex with GTT
To be Published
|
|
4E8E
 
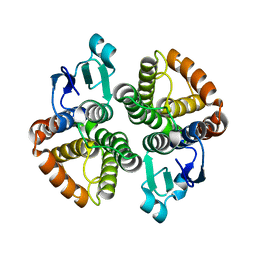 | | Structural characterization of Bombyx mori glutathione transferase BmGSTD1 | | Descriptor: | Glutathione S-transferase | | Authors: | Tan, X, Ma, X.X, Hu, X.M, Chen, Q.M, Zhao, P, Xia, Q.Y, Zhou, C.Z. | | Deposit date: | 2012-03-20 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural characterization of Bombyx mori glutathione transferase BmGSTD1
To be Published
|
|
3N3F
 
 | |
