1XMN
 
 | | Crystal structure of thrombin bound to heparin | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ... | | Authors: | Carter, W.J, Cama, E, Huntington, J.A. | | Deposit date: | 2004-10-04 | | Release date: | 2004-11-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of thrombin bound to heparin
J.Biol.Chem., 280, 2005
|
|
9Q1N
 
 | |
7WQQ
 
 | | Retinoic acid receptor alpha mutant - N299H | | Descriptor: | 4-[(E)-3-(3,5-ditert-butylphenyl)-3-oxidanylidene-prop-1-enyl]benzoic acid, Peptide from Nuclear receptor coactivator 1, Retinoic acid receptor alpha | | Authors: | Huang, X.X, Ng, L.M, Teh, B.T. | | Deposit date: | 2022-01-25 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Effects of breast fibroepithelial tumor associated retinoic acid receptor alpha ligand binding domain mutations on receptor function and retinoid signaling
To Be Published
|
|
7WUP
 
 | | The crystal structure of ApiI | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ApiI, ... | | Authors: | Zhou, J, Lu, J. | | Deposit date: | 2022-02-09 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of ApiI
To Be Published
|
|
7WVS
 
 | | The structure of FinI complex with SAM | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Methyltransf_2 domain-containing protein, ... | | Authors: | Lu, J, Zhou, J. | | Deposit date: | 2022-02-11 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of FinI complex with SAM
To Be Published
|
|
2LXI
 
 | |
9PSE
 
 | | In situ MicroED structure of IL-5 activated human eosinophil major basic protein-1 | | Descriptor: | Bone marrow proteoglycan, CHLORIDE ION | | Authors: | Yang, J.E, Bingman, C.A, Mitchell, J, Mosher, D, Wright, E.R. | | Deposit date: | 2025-07-25 | | Release date: | 2025-09-03 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.18 Å) | | Cite: | In situ microED structure of the Eosinophil major basic protein-1
To Be Published
|
|
7XCT
 
 | | Cryo-EM structure of Dot1L and H2BK34ub-H3K79Nle nucleosome 2:1 complex | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | Deposit date: | 2022-03-25 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
7XIJ
 
 | | Crystal structure of CBP bromodomain liganded with Y08175 | | Descriptor: | 3-[(1-ethanoyl-5-methoxy-indol-3-yl)carbonylamino]-4-fluoranyl-5-(1-methylpyrazol-4-yl)benzoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Xiang, Q, Wang, C, Wu, T, Zhang, Y, Zhang, C, Luo, G, Wu, X, Shen, H, Xu, Y. | | Deposit date: | 2022-04-13 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of CBP bromodomain liganded with Y08175
To Be Published
|
|
4WUV
 
 | | Crystal Structure of a putative D-Mannonate oxidoreductase from Haemophilus influenza (Avi_5165, TARGET EFI-513796) with bound NAD | | Descriptor: | 1,2-ETHANEDIOL, 2-hydroxycyclohexanecarboxyl-CoA dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2014-11-03 | | Release date: | 2014-11-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Crystal structure of a putative D-Mannonate oxidoreductase from Haemophilus influenza (Avi_5165, TARGET EFI-513796) with bound NAD
To be published
|
|
9NKF
 
 | |
9NKJ
 
 | |
9NKG
 
 | |
9NIG
 
 | | PB TCR in complex with HLA-DR4 presenting citrullinated Tenascin C peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dao, H.T, Loh, T.J, Lim, J.J, Rossjohn, J. | | Deposit date: | 2025-02-26 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (3.20008039 Å) | | Cite: | PB TCR in complex with HLA-DR4 presenting citrullinated Tenascin C peptide
To Be Published
|
|
2MA6
 
 | | Solution NMR Structure of the RING finger domain from the Kip1 ubiquitination-promoting E3 complex protein 1 (KPC1/RNF123) from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8700A | | Descriptor: | E3 ubiquitin-protein ligase RNF123, ZINC ION | | Authors: | Ramelot, T.A, Yang, Y, Janjua, H, Kohan, E, Wang, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-28 | | Release date: | 2013-07-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the RING finger domain from the Kip1 ubiquitination-promoting E3 complex protein 1 (KPC1/RNF123) from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8700A
To be Published
|
|
7QTI
 
 | | SARS-CoV-2 S Omicron Spike B.1.1.529 - 3-P2G3 and 1-P5C3 Fabs (Global) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, P2G3 Heavy Chain, ... | | Authors: | Ni, D, Lau, K, Turelli, P, Fenwick, C, Perez, L, Pojer, F, Stahlberg, H, Pantaleo, G, Trono, D. | | Deposit date: | 2022-01-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Patient-derived monoclonal antibody neutralizes SARS-CoV-2 Omicron variants and confers full protection in monkeys.
Nat Microbiol, 7, 2022
|
|
7WUG
 
 | | GID subcomplex: Gid12 bound Substrate Receptor Scaffolding module | | Descriptor: | Glucose-induced degradation protein 8, HLJ1_G0042170.mRNA.1.CDS.1, Vacuolar import and degradation protein 24, ... | | Authors: | Qiao, S, Cheng, J.D, Schulman, B.A. | | Deposit date: | 2022-02-08 | | Release date: | 2022-06-08 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of Gid12-bound GID E3 reveal steric blockade as a mechanism inhibiting substrate ubiquitylation.
Nat Commun, 13, 2022
|
|
2M0D
 
 | |
7W7E
 
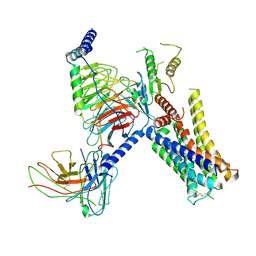 | | Cryo-EM structure of the alpha2A adrenergic receptor GoA signaling complex bound to a biased agonist | | Descriptor: | 5-(3-bicyclo[4.2.0]octa-1,3,5-trienyl)-1,2,3,6-tetrahydropyridine, Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, J, Fink, E.A, Shoichet, B.K, Du, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-based discovery of nonopioid analgesics acting through the alpha 2A -adrenergic receptor.
Science, 377, 2022
|
|
2M0Z
 
 | | cis form of a photoswitchable PDZ domain crosslinked with an azobenzene derivative | | Descriptor: | 3,3'-(E)-diazene-1,2-diylbis{6-[(chloroacetyl)amino]benzenesulfonic acid}, Tyrosine-protein phosphatase non-receptor type 13 | | Authors: | Walser, R, Zerbe, O, Hamm, P. | | Deposit date: | 2012-11-09 | | Release date: | 2013-07-03 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Kinetic response of a photoperturbed allosteric protein.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6CUF
 
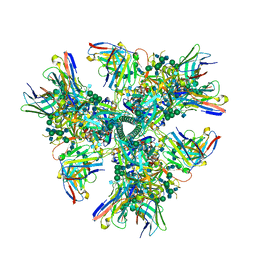 | | Cryo-EM structure at 4.2 A resolution of vaccine-elicited antibody vFP1.01 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Acharya, P, Carragher, B, Potter, C.S, Kwong, P.D. | | Deposit date: | 2018-03-26 | | Release date: | 2018-07-25 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Complete functional mapping of infection- and vaccine-elicited antibodies against the fusion peptide of HIV.
PLoS Pathog., 14, 2018
|
|
9O9V
 
 | |
9OG5
 
 | | SARS-COV-2-6P-MUT7 S PROTEIN-DY-III-281 complex 1 RBD up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Chandravanshi, M, Niu, L, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2025-04-30 | | Release date: | 2025-10-01 | | Last modified: | 2025-10-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Optimization of VE607 to generate analogs with improved neutralization activities against SARS-CoV-2 variants.
J.Virol., 2025
|
|
9O2Q
 
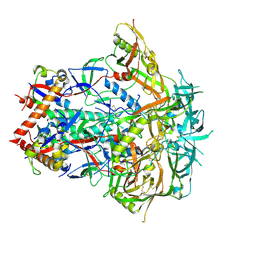 | | BG505-DS SOSIP in complex with 007 bNAb Fabs - Class 0 (unbound) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | DeLaitsch, A.T, Bjorkman, P.J. | | Deposit date: | 2025-04-04 | | Release date: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Identification of a broad and potent V3 glycan site bNAb targeting an N332 gp120 glycan-independent epitope.
Biorxiv, 2025
|
|
7XCR
 
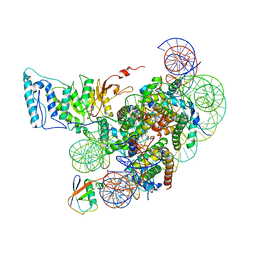 | | Cryo-EM structure of Dot1L and H2BK34ub-H3K79Nle nucleosome 1:1 complex | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | Deposit date: | 2022-03-25 | | Release date: | 2022-04-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
