6D49
 
 | | Cell Surface Receptor in Complex with Ligand at 1.80-A Resolution | | Descriptor: | 2-aminoethyl 5-{[(4-cyclohexyl-1H-1,2,3-triazol-1-yl)acetyl]amino}-3,5,9-trideoxy-9-[(4-hydroxy-3,5-dimethylbenzene-1-carbonyl)amino]-D-glycero-alpha-D-galacto-non-2-ulopyranonosyl-(2->6)-beta-D-galactopyranosyl-(1->4)-beta-D-glucopyranoside, GLYCEROL, Myeloid cell surface antigen CD33 | | Authors: | Hermans, S.J, Miles, L.A, Parker, M.W. | | Deposit date: | 2018-04-17 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Small Molecule Binding to Alzheimer Risk Factor CD33 Promotes A beta Phagocytosis.
Iscience, 19, 2019
|
|
5EVK
 
 | | Crystal structure of the metallo-beta-lactamase L1 in complex with the bisthiazolidine inhibitor L-CS319 | | Descriptor: | (3R,5R,7aS)-5-(sulfanylmethyl)tetrahydro[1,3]thiazolo[4,3-b][1,3]thiazole-3-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2015-11-19 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.627 Å) | | Cite: | Cross-class metallo-beta-lactamase inhibition by bisthiazolidines reveals multiple binding modes.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EYR
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound 5 at 1.57A resolution | | Descriptor: | 3-[1-[4-(methylaminomethyl)phenyl]piperidin-4-yl]-1-pyrrolidin-1-yl-propan-1-one, EthR | | Authors: | Surade, S, Blaszczyk, M, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2015-11-25 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A fragment merging approach towards the development of small molecule inhibitors of Mycobacterium tuberculosis EthR for use as ethionamide boosters.
Org.Biomol.Chem., 14, 2016
|
|
9OUG
 
 | | Influenza A Virus Nucleoprotein(8-498)NP complex with rac-5-(4-(((2R,6R)-6-(methoxymethyl)-6-methyl-1,4-dioxan-2-yl)methoxy)phenyl)-2-oxo-6-(trifluoromethyl)-1,2-dihydropyridine-3-carboxamide (Compound 20) | | Descriptor: | 5-(4-{[(2R,6S)-6-(methoxymethyl)-6-methyl-1,4-dioxan-2-yl]methoxy}phenyl)-2-oxo-6-(trifluoromethyl)-1,2-dihydropyridine-3-carboxamide, Nucleoprotein | | Authors: | Mamo, M. | | Deposit date: | 2025-05-28 | | Release date: | 2025-08-06 | | Last modified: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery and Optimization of a Novel Series of Influenza A Virus Replication Inhibitors Targeting the Nucleoprotein Protein-Protein Interaction.
J.Med.Chem., 68, 2025
|
|
6CX9
 
 | | Structure of alpha-GSA[16,6P] bound by CD1d and in complex with the Va14Vb8.2 TCR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Wang, J, Zajonc, D. | | Deposit date: | 2018-04-02 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
6VEO
 
 | | ATAD2B bromodomain in complex with 4-({[(3R,4R)-4-{[3-methyl-5-(5-methylpyridin-3-yl)-2-oxo-1,2-dihydro-1,7-naphthyridin-8-yl]amino}piperidin-3-yl]oxy}methyl)-1lambda~6~-thiane-1,1-dione (compound 38) | | Descriptor: | 4-({[(3R,4R)-4-{[3-methyl-5-(5-methylpyridin-3-yl)-2-oxo-1,2-dihydro-1,7-naphthyridin-8-yl]amino}piperidin-3-yl]oxy}methyl)-1lambda~6~-thiane-1,1-dione, ATPase family AAA domain-containing protein 2B, SULFATE ION | | Authors: | Glass, K.C. | | Deposit date: | 2020-01-02 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into the Recognition of Mono- and Diacetylated Histones by the ATAD2B Bromodomain.
J.Med.Chem., 63, 2020
|
|
7REK
 
 | |
8CRE
 
 | | Crystal structure of the Candida albicans 80S ribosome in complex with geneticin G418 | | Descriptor: | 18S, 25S, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Kolosova, O, Zgadzay, Y, Yusupov, M. | | Deposit date: | 2023-03-08 | | Release date: | 2024-09-18 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanism of read-through enhancement by aminoglycosides and mefloquine
Proc.Natl.Acad.Sci.USA, 2025
|
|
1XHB
 
 | | The Crystal Structure of UDP-GalNAc: polypeptide alpha-N-acetylgalactosaminyltransferase-T1 | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, Polypeptide N-acetylgalactosaminyltransferase 1, ... | | Authors: | Fritz, T.A, Hurley, J.H, Trinh, L.B, Shiloach, J, Tabak, L.A. | | Deposit date: | 2004-09-17 | | Release date: | 2004-10-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The beginnings of mucin biosynthesis: The crystal structure of UDP-GalNAc:polypeptide {alpha}-N-acetylgalactosaminyltransferase-T1
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6VFP
 
 | | Crystal structure of human protocadherin 1 EC1-EC4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin-1, ... | | Authors: | Brasch, J, Harrison, O.J, Shapiro, L. | | Deposit date: | 2020-01-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Family-wide Structural and Biophysical Analysis of Binding Interactions among Non-clustered delta-Protocadherins.
Cell Rep, 30, 2020
|
|
8CQ7
 
 | | Crystal structure of phyllanthoside bound to the Candida albicans 80S ribosome | | Descriptor: | 18S, 25S, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Kolosova, O, Zgadzay, Y, Yusupov, M. | | Deposit date: | 2023-03-03 | | Release date: | 2024-09-11 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism of read-through enhancement by aminoglycosides and mefloquine
Proc.Natl.Acad.Sci.USA, 2025
|
|
6D4A
 
 | | Cell Surface Receptor with Bound Ligand at 1.75-A Resolution | | Descriptor: | 2-aminoethyl 5-{[(4-cyclohexyl-1H-1,2,3-triazol-1-yl)acetyl]amino}-3,5,9-trideoxy-9-[(4-hydroxy-3,5-dimethylbenzene-1-carbonyl)amino]-D-glycero-alpha-D-galacto-non-2-ulopyranonosyl-(2->6)-beta-D-galactopyranosyl-(1->4)-beta-D-glucopyranoside, GLYCEROL, Myeloid cell surface antigen CD33 | | Authors: | Hermans, S.J, Miles, L.A, Parker, M.W. | | Deposit date: | 2018-04-17 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Small Molecule Binding to Alzheimer Risk Factor CD33 Promotes A beta Phagocytosis.
Iscience, 19, 2019
|
|
5EXN
 
 | | FACTOR XIA (C500S [C122S]) IN COMPLEX WITH THE INHIBITOR methyl ~{N}-[4-[2-[(1~{S})-1-[[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]amino]-2-phenyl-ethyl]pyridin-4-yl]phenyl]carbamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XIa light chain, methyl ~{N}-[4-[2-[(1~{S})-1-[[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]amino]-2-phenyl-ethyl]pyridin-4-yl]phenyl]carbamate | | Authors: | Sheriff, S. | | Deposit date: | 2015-11-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Orally bioavailable pyridine and pyrimidine-based Factor XIa inhibitors: Discovery of the methyl N-phenyl carbamate P2 prime group
Bioorg.Med.Chem., 24, 2016
|
|
7WCH
 
 | |
9PLN
 
 | | Locally-refined structure of alpha2a adrenergic receptor in complex with Go heterotrimer, scFv16, and N-(5-methylnaphthalen-1-yl)pyridin-4-amine (compound 4905) | | Descriptor: | Alpha-2A adrenergic receptor, N-(5-methylnaphthalen-1-yl)pyridin-4-amine | | Authors: | Srinivasan, K, Xu, X, Mailhot, O, Manglik, A, Shoichet, B. | | Deposit date: | 2025-07-15 | | Release date: | 2025-08-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Toward a Random Background for Ligand Optimization
To Be Published
|
|
3L4U
 
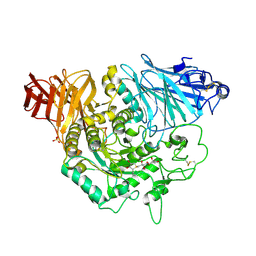 | | Crystal complex of N-terminal Human Maltase-Glucoamylase with de-O-sulfonated kotalanol | | Descriptor: | (2R,3S,4S)-1-[(2S,3S,4R,5R,6S)-2,3,4,5,6,7-hexahydroxyheptyl]-3,4-dihydroxy-2-(hydroxymethyl)tetrahydrothiophenium (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New glucosidase inhibitors from an ayurvedic herbal treatment for type 2 diabetes: structures and inhibition of human intestinal maltase-glucoamylase with compounds from Salacia reticulata.
Biochemistry, 49, 2010
|
|
9NXJ
 
 | | The GH43 domain of an alpha-l-arabinofuranosidase (AtAbf43C_GH43) from Acetivibrio thermocellus DSM1313 | | Descriptor: | Glycoside hydrolase family 43, SULFATE ION, alpha-L-arabinofuranose | | Authors: | Galindo, J.L, Jeffrey, P.D, Conway, J.M. | | Deposit date: | 2025-03-25 | | Release date: | 2025-08-20 | | Last modified: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Functional and structural characterization of AtAbf43C: an exo-1,5-alpha-L-arabinofuranosidase from Acetivibrio thermocellus DSM1313.
Biochem.J., 482, 2025
|
|
9OND
 
 | |
5F1J
 
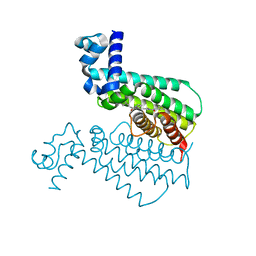 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound 1 at 1.63A resolution | | Descriptor: | 3-cyclopentyl-1-pyrrolidin-1-yl-propan-1-one, HTH-type transcriptional regulator EthR | | Authors: | Surade, S, Blaszczyk, M, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2015-11-30 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | A fragment merging approach towards the development of small molecule inhibitors of Mycobacterium tuberculosis EthR for use as ethionamide boosters.
Org.Biomol.Chem., 14, 2016
|
|
7WH8
 
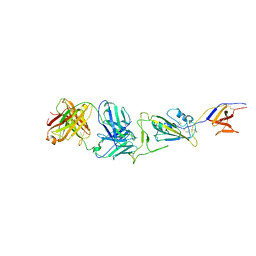 | |
9ON7
 
 | |
6V61
 
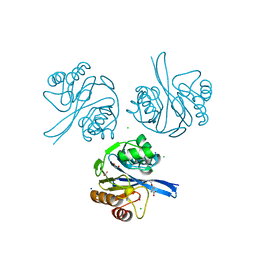 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in the Complex with the Inhibitor Captopril | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-04 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in the Complex with the Inhibitor Captopril.
To Be Published
|
|
6D05
 
 | | Cryo-EM structure of a Plasmodium vivax invasion complex essential for entry into human reticulocytes; two molecules of parasite ligand, subclass 2. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Gruszczyk, J, Huang, R.K, Hong, C, Yu, Z, Tham, W.H. | | Deposit date: | 2018-04-10 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
7RN5
 
 | |
9OJX
 
 | | Crystal structure of E. coli ApaH in complex with GDP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bis(5'-nucleosyl)-tetraphosphatase [symmetrical], GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nuthanakanti, A, Serganov, A. | | Deposit date: | 2025-05-08 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | ApaH decaps Np 4 N-capped RNAs in two alternative orientations.
Nat.Chem.Biol., 2025
|
|
