3D1I
 
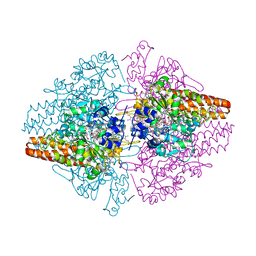 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with nitrite | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2008-05-06 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens
J.Mol.Biol., 389, 2009
|
|
1B7N
 
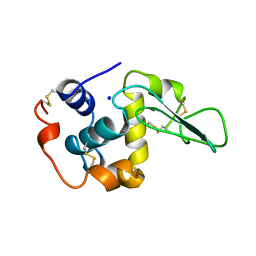 | | VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Ota, M, Ogasahara, K, Yamagata, Y, Nishikawa, K, Yutani, K. | | Deposit date: | 1999-01-24 | | Release date: | 1999-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental verification of the 'stability profile of mutant protein' (SPMP) data using mutant human lysozymes.
Protein Eng., 12, 1999
|
|
1B5V
 
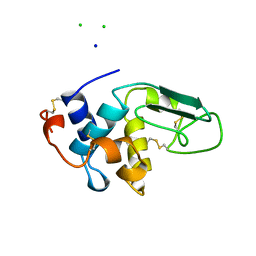 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS | | Descriptor: | CHLORIDE ION, PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
1B7L
 
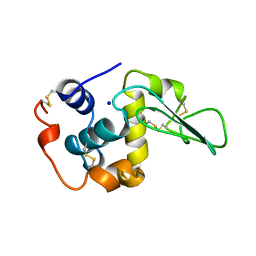 | | VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Ota, M, Ogasahara, K, Yamagata, Y, Nishikawa, K, Yutani, K. | | Deposit date: | 1999-01-24 | | Release date: | 1999-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental verification of the 'stability profile of mutant protein' (SPMP) data using mutant human lysozymes.
Protein Eng., 12, 1999
|
|
1B5Y
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
5HXI
 
 | | 2-Methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5HN bound | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxypyridine-3-carboxylic acid, BETA-MERCAPTOETHANOL, ... | | Authors: | Kobayashi, J, Mikami, B. | | Deposit date: | 2016-01-30 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Role of the Tyr270 residue in 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase from Mesorhizobium loti
J. Biosci. Bioeng., 123, 2017
|
|
5HHH
 
 | | Structure of human DNA polymerase beta Host-Guest complexed with the control G for N7-CBZ-platination | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*AP*GP*GP*AP*GP*CP*AP*GP*G)-3'), DNA (5'-D(P*CP*CP*TP*GP*CP*TP*CP*CP*TP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Koag, M.-C, Lee, S. | | Deposit date: | 2016-01-11 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.363 Å) | | Cite: | Synthesis, structure, and biological evaluation of a platinum-carbazole conjugate.
Chem Biol Drug Des, 91, 2018
|
|
3FF1
 
 | | Structure of Glucose 6-phosphate Isomerase from Staphylococcus aureus | | Descriptor: | GLUCOSE-6-PHOSPHATE, Glucose-6-phosphate isomerase, SODIUM ION | | Authors: | Anderson, S.M, Brunzelle, J.S, Onopriyenko, O, Peterson, S, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-12-01 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Glucose 6-phosphate Isomerase from Staphylococcus aureus
TO BE PUBLISHED
|
|
8W6C
 
 | | CryoEM structure of NaDC1 with Citrate | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, CHOLESTEROL HEMISUCCINATE, CITRIC ACID, ... | | Authors: | Chi, X, Chen, Y, Li, Y, Dai, L, Zhang, Y, Shen, Y, Chen, Y, Shi, T, Yang, H, Wang, Z, Yan, R. | | Deposit date: | 2023-08-28 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
8W6H
 
 | | NaS1 with sulfate - IN/IN state | | Descriptor: | CHOLESTEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Chi, X, Chen, Y, Li, Y, Zhang, Y, Shen, Y, Chen, Y, Wang, Z, Yan, R. | | Deposit date: | 2023-08-28 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
3D7R
 
 | | Crystal structure of a putative esterase from Staphylococcus aureus | | Descriptor: | BROMIDE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Lam, R, Battaile, K.P, Chan, T, Romanov, V, Lam, K, Soloveychik, M, Wu-Brown, J, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2008-05-21 | | Release date: | 2009-06-09 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of a putative esterase from Staphylococcus aureus
To be Published
|
|
5HXA
 
 | |
7OLB
 
 | | Crystal structure of Pab-AGOG, an 8-oxoguanine DNA glycosylase from Pyrococcus abyssi | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-05-19 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
1B7O
 
 | | VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Ota, M, Ogasahara, K, Yamagata, Y, Nishikawa, K, Yutani, K. | | Deposit date: | 1999-01-25 | | Release date: | 1999-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental verification of the 'stability profile of mutant protein' (SPMP) data using mutant human lysozymes.
Protein Eng., 12, 1999
|
|
3G7R
 
 | | Crystal structure of SCO4454, a TetR-family transcriptional regulator from Streptomyces coelicolor | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Xu, X, Chang, C, Gu, J, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-10 | | Release date: | 2009-03-10 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure and ligand specificity of SCO4454, a TetR-family transcriptional regulator from Streptomyces coelicolor
To be Published
|
|
3DIM
 
 | |
3DC7
 
 | | Crystal structure of the protein Q88SR8 from Lactobacillus plantarum. Northeast Structural Genomics consortium target LpR109. | | Descriptor: | MAGNESIUM ION, Putative uncharacterized protein lp_3323, SODIUM ION, ... | | Authors: | Kuzin, A.P, Abashidze, M, Vorobiev, S.M, Seetharaman, J, Zhao, L, Mao, L, Ciccosanti, C, Xiao, R, Nair, R, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-03 | | Release date: | 2008-08-05 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of the protein Q88SR8 from Lactobacillus plantarum. Northeast Structural Genomics consortium target LpR109. (CASP Target)
To be Published
|
|
5F8A
 
 | | Crystal structure of the ternary EcoRV-DNA-Lu complex with uncleaved DNA substrate. Lanthanide binding to EcoRV-DNA complex inhibits cleavage. | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*TP*CP*TP*TP*T)-3'), LUTETIUM (III) ION, ... | | Authors: | Sangani, S.S, Kehr, A.D, Sinha, K, Rule, G.S, Jen-Jacobson, L. | | Deposit date: | 2015-12-09 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Metal Ion Binding at the Catalytic Site Induces Widely Distributed Changes in a Sequence Specific Protein-DNA Complex.
Biochemistry, 55, 2016
|
|
7OYL
 
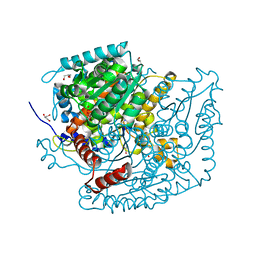 | | Phosphoglucose isomerase of Aspergillus fumigatus in complexed with Glucose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Raimi, O.G, Yan, K, Fang, W, van Aalten, D.M.F. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Phosphoglucose Isomerase Is Important for Aspergillus fumigatus Cell Wall Biogenesis.
Mbio, 13, 2022
|
|
7P0W
 
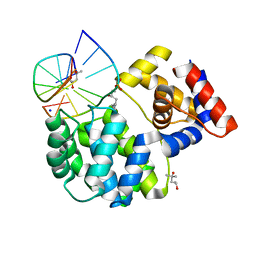 | | Crystal structure of a trapped Pab-AGOG/double-standed DNA covalent intermediate (DNA containing thymine opposite to lesion) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*(PED)P*TP*TP*TP*CP*T)-3'), ... | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
3T46
 
 | |
3MCQ
 
 | |
3Q2H
 
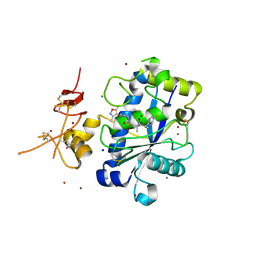 | | Adamts1 in complex with N-hydroxyformamide inhibitors of ADAM-TS4 | | Descriptor: | A disintegrin and metalloproteinase with thrombospondin motifs 1, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The design and synthesis of novel N-hydroxyformamide inhibitors of ADAM-TS4 for the treatment of osteoarthritis
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5HK7
 
 | | Bacterial sodium channel pore, 2.95 Angstrom resolution | | Descriptor: | 2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}ethyl heptadecanoate, CHLORIDE ION, Ion transport protein, ... | | Authors: | Shaya, D, Findeisen, F, Rohaim, A, Minor, D.L. | | Deposit date: | 2016-01-14 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Unfolding of a Temperature-Sensitive Domain Controls Voltage-Gated Channel Activation.
Cell, 164, 2016
|
|
3DT0
 
 | | Understanding Thrombin Inhibition | | Descriptor: | Hirudin variant-1, N-(3-chlorobenzyl)-1-(4-methylpentanoyl)-L-prolinamide, SODIUM ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-07-14 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Understanding Thrombin Inhibition
To be Published
|
|
