8SWS
 
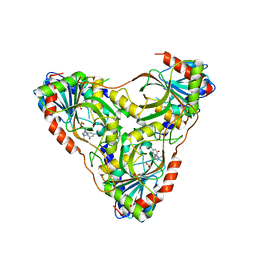 | | Structure of K. lactis PNP S42E-H98R variant bound to transition state analog DADMe-IMMUCILLIN G and sulfate | | Descriptor: | 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Fedorov, E, Ghosh, A. | | Deposit date: | 2023-05-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Phosphate Binding in PNP Alters Transition-State Analogue Affinity and Subunit Cooperativity.
Biochemistry, 62, 2023
|
|
8SWR
 
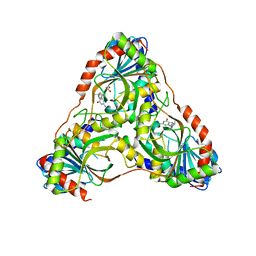 | | Structure of K. lactis PNP S42E variant bound to transition state analog DADMe-IMMUCILLIN G and sulfate | | Descriptor: | 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, GUANINE, Purine nucleoside phosphorylase, ... | | Authors: | Fedorov, E, Ghosh, A. | | Deposit date: | 2023-05-19 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Phosphate Binding in PNP Alters Transition-State Analogue Affinity and Subunit Cooperativity.
Biochemistry, 62, 2023
|
|
8SWQ
 
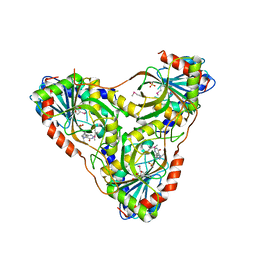 | | Structure of K. lactis PNP bound to transition state analog DADMe-IMMUCILLIN H and sulfate | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, GLYCEROL, Purine nucleoside phosphorylase, ... | | Authors: | Fedorov, E, Ghosh, A. | | Deposit date: | 2023-05-19 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Phosphate Binding in PNP Alters Transition-State Analogue Affinity and Subunit Cooperativity.
Biochemistry, 62, 2023
|
|
8SWP
 
 | | Structure of K. lactis PNP bound to hypoxanthine | | Descriptor: | ACETATE ION, HYPOXANTHINE, Purine nucleoside phosphorylase | | Authors: | Fedorov, E, Ghosh, A. | | Deposit date: | 2023-05-19 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphate Binding in PNP Alters Transition-State Analogue Affinity and Subunit Cooperativity.
Biochemistry, 62, 2023
|
|
8SWO
 
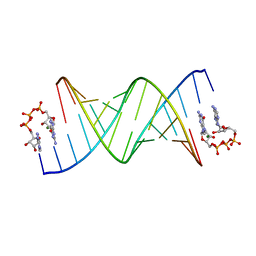 | | GpppA dinucleotide ligand binding to RNA UC template | | Descriptor: | GUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, MAGNESIUM ION, RNA (5'-R(*(TLN)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*G*(GA3))-3') | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2023-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Insight into the structures of unusual base pairs in RNA complexes containing a primer/template/adenosine ligand.
Rsc Chem Biol, 4, 2023
|
|
8SWN
 
 | |
8SWM
 
 | |
8SWL
 
 | |
8SWK
 
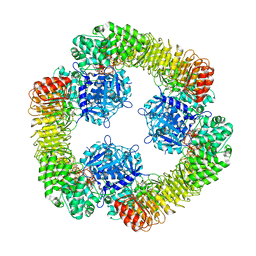 | | Cryo-EM structure of NLRP3 closed hexamer | | Descriptor: | 1-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-3-(1,2,3,5-tetrahydro-s-indacen-4-yl)urea, ADENOSINE-5'-TRIPHOSPHATE, NACHT, ... | | Authors: | Yu, X, Matico, R.E, Miller, R, Schoubroeck, B.V, Grauwen, K, Suarez, J, Pietrak, B, Haloi, N, Yin, Y, Tresadern, G.J, Perez-Benito, L, Lindahl, E, Bottelbergs, A, Oehlrich, D, Opdenbosch, N.V, Sharma, S. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Cryo-EM structures of NLRP3 reveal its self-activation mechanism
Nat Commun, 2024
|
|
8SWJ
 
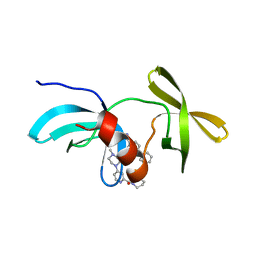 | | Co-crystal structure of 53BP1 tandem Tudor domains in complex with UNC8531 | | Descriptor: | (3S)-N-(4'-carbamoyl[1,1'-biphenyl]-3-yl)-1-[4-(4-methylpiperazin-1-yl)pyridine-2-carbonyl]piperidine-3-carboxamide, TP53-binding protein 1, UNKNOWN ATOM OR ION | | Authors: | Zeng, H, Dong, A, Shell, D.J, Foley, C, Pearce, K.H, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Co-crystal structure of 53BP1 tandem Tudor domains in complex with UNC8531
To be published
|
|
8SWI
 
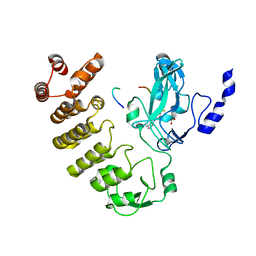 | |
8SWH
 
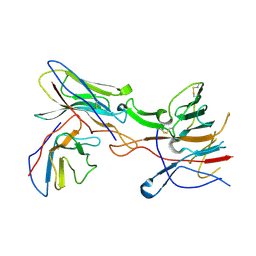 | |
8SWG
 
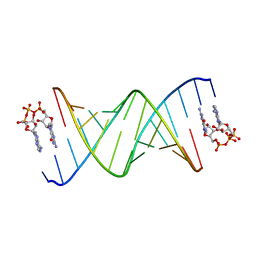 | | RNA duplex bound with GpppA dinucleotide ligand | | Descriptor: | GUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*G*(GA3))-3') | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2023-05-18 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insight into the structures of unusual base pairs in RNA complexes containing a primer/template/adenosine ligand.
Rsc Chem Biol, 4, 2023
|
|
8SWF
 
 | | Cryo-EM structure of NLRP3 open octamer | | Descriptor: | NACHT, LRR and PYD domains-containing protein 3 | | Authors: | Yu, X, Matico, R.E, Miller, R, Schoubroeck, B.V, Grauwen, K, Suarez, J, Pietrak, B, Haloi, N, Yin, Y, Tresadern, G.J, Perez-Benito, L, Lindahl, E, Bottelbergs, A, Oehlrich, D, Opdenbosch, N.V, Sharma, S. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Cryo-EM structures of NLRP3 reveal its self-activation mechanism
Nat Commun, 2024
|
|
8SWE
 
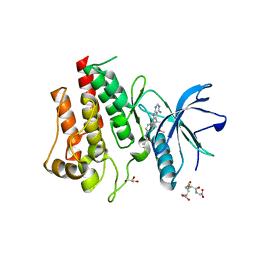 | | FGFR2 Kinase Domain Bound to Reversible Inhibitor Cmpd 3 | | Descriptor: | Fibroblast growth factor receptor 2, GLUTATHIONE, GLYCEROL, ... | | Authors: | Valverde, R, Foster, L. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Discovery of lirafugratinib (RLY-4008), a highly selective irreversible small-molecule inhibitor of FGFR2.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SWD
 
 | |
8SWC
 
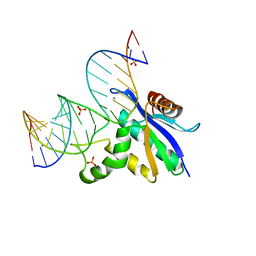 | |
8SWB
 
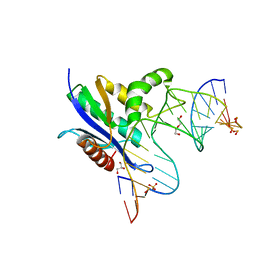 | | RNase H complex with streopure ASO and RNA | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA (5'-R(*(OMC)P*(N7X)P*(T39)P*(C5L)P*(A2M))-D(P*(SC)P*(PST)P*(SC)P*AP*(SC)P*(SC)P*(RC)P*(AS)P*(SC)P*(PST))-R(P*(6OO)P*(RFJ)P*(6OO)P*(6OO)P*(6NW))-3'), GLYCEROL, ... | | Authors: | Cho, Y.-J, Iwamoto, N. | | Deposit date: | 2023-05-18 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of RNase H/RNA/PS-ASO complex at an atomic level
To Be Published
|
|
8SWA
 
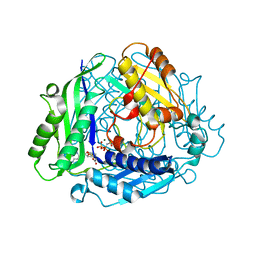 | | Crystal structure of the human S-adenosylmethionine synthetase 1 in complex with SAM and PPNP | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Bruemmer, K.J, Pham, V.N, Toh, J.D.W. | | Deposit date: | 2023-05-18 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Formaldehyde regulates S -adenosylmethionine biosynthesis and one-carbon metabolism.
Science, 382, 2023
|
|
8SW9
 
 | | Plasmodium falciparum M17 (A460S) mutant | | Descriptor: | CARBONATE ION, Leucine aminopeptidase, PENTAETHYLENE GLYCOL, ... | | Authors: | McGowan, S, Suraweera, C, Drinkwater, N. | | Deposit date: | 2023-05-17 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Characterisation of a novel antimalarial agent targeting haemaglobin digestion that shows cross-species reactivity and excellent in vivo properties.
Mbio, 2024
|
|
8SW8
 
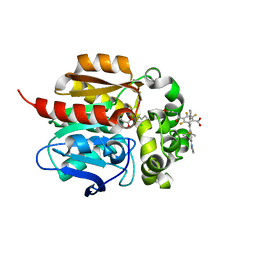 | | Crystal Structure of HaloTag7 bound to JF669-HaloTag ligand | | Descriptor: | 1-[(4aM,10P)-7-(azetidin-1-yl)-10-[2-carboxy-3,4,6-trifluoro-5-({2-[2-(hexyloxy)ethoxy]ethyl}carbamoyl)phenyl]-5,5-dimethyldibenzo[b,e]silin-3(5H)-ylidene]azetidin-1-ium, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Farrants, H, Schreiter, E.R. | | Deposit date: | 2023-05-17 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A modular chemigenetic calcium indicator for multiplexed in vivo functional imaging
To Be Published
|
|
8SW7
 
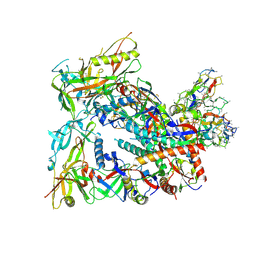 | | BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody FP1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 Boost 2 gp120, BG505 Boost 2 gp41, ... | | Authors: | Pratap, P.P, Antansijevic, A, Ozorowski, G, Ward, A.B. | | Deposit date: | 2023-05-17 | | Release date: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Focusing antibody responses to the fusion peptide in rhesus macaques.
Biorxiv, 2023
|
|
8SW6
 
 | |
8SW5
 
 | |
8SW4
 
 | | BG505 GT1.1 SOSIP in complex with NHP Fabs 21N13, 21M20 and RM20A3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 21M20 heavy chain variable region, ... | | Authors: | Ozorowski, G, Torres, J.L, Zhang, S, Ward, A.B. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Neutralizing antibodies induced in non-human primates by germline targeting Env trimer
To Be Published
|
|
