5N0H
 
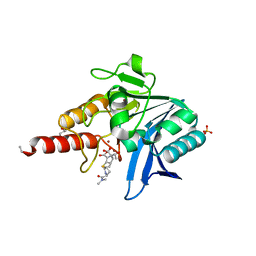 | | Crystal structure of NDM-1 in complex with hydrolyzed meropenem - new refinement | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, King, D.T, Strynadka, N.C.J. | | Deposit date: | 2017-02-03 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
8X4M
 
 | | Crystal structure of the glycosyltransferase domain of Legionella SetA in complex with UPD | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Im, H.N, Lee, Y, Jang, D.M, Hahn, H, Kim, H.S. | | Deposit date: | 2023-11-15 | | Release date: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of the glycosyltransferase domain of Legionella SetA in complex with UPD
To Be Published
|
|
6V8D
 
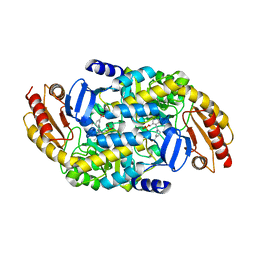 | | Design, Synthesis, and Mechanism of Fluorine-substituted Cyclohexene Analogues of GAMA-Aminobutyric Acid (GABA) as Selective Ornithine Aminotransferase Inactivators | | Descriptor: | (3Z)-3-iminocyclohex-1-ene-1-carboxylic acid, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Zhu, W, Doubleday, P.T, Catlin, D.S, Weerawarna, P, Butrin, A, Shen, S, Kelleher, N.L, Liu, D, Silverman, R.B. | | Deposit date: | 2019-12-10 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design, Synthesis, and Mechanism of Fluorine-substituted Cyclohexene Analogues of GAMA-Aminobutyric Acid (GABA) as Selective Ornithine Aminotransferase Inactivators
To Be Published
|
|
7L73
 
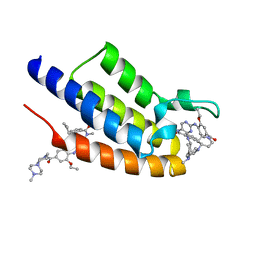 | | Crystal structure of the first bromodomain (BD1) of human BRDT bound to ERK5-IN-1 | | Descriptor: | 11-cyclopentyl-2-({2-ethoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain testis-specific protein | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2020-12-25 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
3WDR
 
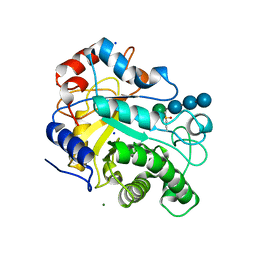 | | Crystal structure of beta-mannanase from a symbiotic protist of the termite Reticulitermes speratus complexed with gluco-manno-oligosaccharide | | Descriptor: | BICARBONATE ION, Beta-mannanase, MAGNESIUM ION, ... | | Authors: | Tsukagoshi, H, Ishida, T, Touhara, K.K, Igarashi, K, Samejima, M, Fushinobu, S, Kitamoto, K, Arioka, M. | | Deposit date: | 2013-06-20 | | Release date: | 2014-03-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Biochemical Analyses of Glycoside Hydrolase Family 26 beta-Mannanase from a Symbiotic Protist of the Termite Reticulitermes speratus
J.Biol.Chem., 289, 2014
|
|
7VDU
 
 | | The structure of cyclin-dependent kinase 2 (CDK2) in complex with Compound 1 | | Descriptor: | Cyclin-dependent kinase 2, [1-[3-fluoranyl-4-[(2-piperidin-4-yloxy-1,6-naphthyridin-7-yl)amino]phenyl]pyrazol-3-yl]methanol | | Authors: | Malojcic, G, Clugston, S.L, Daniels, M, Harmange, J.C, Ledeborer, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Discovery and Optimization of Highly Selective Inhibitors of CDK5.
J.Med.Chem., 65, 2022
|
|
7L9A
 
 | | Crystal structure of BRDT bromodomain 2 in complex with CDD-1102 | | Descriptor: | BETA-MERCAPTOETHANOL, Bromodomain testis-specific protein, N~1~-(5-{[3-(4-amino-2-methylphenyl)-1-methyl-1H-indazole-5-carbonyl]amino}-2-methylphenyl)-N~4~-methylbenzene-1,4-dicarboxamide | | Authors: | Sharma, R, Kaur, G, Yu, Z, Ku, A.F, Anglin, J.L, Ucisik, M.N, Faver, J.C, Sankaran, B, Kim, C, Matzuk, M.M. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery and characterization of bromodomain 2-specific inhibitors of BRDT.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4UCD
 
 | | N-terminal globular domain of the RSV Nucleoprotein in complex with the Nucleoprotein Phosphoprotein interaction inhibitor M81 | | Descriptor: | 1-[(2-chlorophenyl)methyl]pyrazole-3,5-dicarboxylic acid, NUCLEOPROTEIN, SULFATE ION | | Authors: | Ouizougun-Oubari, M, Pereira, N, Tarus, B, Galloux, M, Tortorici, M.-A, Hoos, S, Baron, B, England, P, Bontems, F, Rey, F.A, Eleouet, J.-F, Sizun, C, Slama-Schwok, A, Duquerroy, S. | | Deposit date: | 2014-12-03 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of the Human Respiratory Syncytial Virus.
J.Virol., 89, 2015
|
|
8Q0Y
 
 | | Endothiapepsin in complex with ligand (3R,5R)-5-(3-(4-bromophenyl)-1,2,4-oxadiazol-5-yl)-3-(2-((methyl(prop-2-yn-1-yl)amino)methyl)thiazol-4-yl)pyrrolidin-3-ol (CBWS-SE-085.1) | | Descriptor: | (3~{R},5~{R})-5-[3-(4-bromophenyl)-1,2,4-oxadiazol-5-yl]-3-[2-[[methyl(prop-2-ynyl)amino]methyl]-1,3-thiazol-4-yl]pyrrolidin-3-ol, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Mueller, J.M, Eckelt, S, Klebe, G, Glinca, S. | | Deposit date: | 2023-07-29 | | Release date: | 2024-08-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Crystal structures of Endothiapepsin with ligands derived from merged fragment hits
To Be Published
|
|
9EWD
 
 | | DNA Polymerase Lambda I493K E529D, TMP:At Ca2+ Ground State Ternary Complex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA polymerase lambda, ... | | Authors: | Nourisson, A, Haouz, A, Missoury, S, Delarue, M. | | Deposit date: | 2024-04-03 | | Release date: | 2025-07-23 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Fidelity, specialization, and evolution of Paramecium PolX DNA polymerases involved in programmed double-strand break DNA repair.
Nucleic Acids Res., 53, 2025
|
|
4Z9A
 
 | | Crystal structure of Low Molecular Weight Protein Tyrosine Phosphatase isoform A complexed with phenylmethanesulfonic acid | | Descriptor: | GLYCEROL, Low molecular weight phosphotyrosine protein phosphatase, SULFATE ION, ... | | Authors: | Trivella, D.B.B, Fonseca, E.M.B, Scorsato, V, Dias, M.P, Aparicio, R. | | Deposit date: | 2015-04-10 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the apo form and a complex of human LMW-PTP with a phosphonic acid provide new evidence of a secondary site potentially related to the anchorage of natural substrates.
Bioorg.Med.Chem., 23, 2015
|
|
3BZI
 
 | | Molecular and structural basis of polo-like kinase 1 substrate recognition: Implications in centrosomal localization | | Descriptor: | 9 mer peptide from M-phase inducer phosphatase 3, FORMIC ACID, Serine/threonine-protein kinase PLK1 | | Authors: | Garcia-Alvarez, B, de Carcer, G, Ibanez, S, Bragado-Nilsson, E, Montoya, G. | | Deposit date: | 2008-01-18 | | Release date: | 2008-02-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular and structural basis of polo-like kinase 1 substrate recognition: Implications in centrosomal localization.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6IJR
 
 | | Human PPARgamma ligand binding domain complexed with SB1495 | | Descriptor: | 16 mer peptide from Nuclear receptor coactivator 1, N-{[3-({[(1S,2R)-2-{[(2E)-2-cyano-4,4-dimethylpent-2-enoyl]amino}cyclopentyl]oxy}methyl)phenyl]methyl}-4-[(4-methylpiperazin-1-yl)methyl]benzamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y, Han, B.W. | | Deposit date: | 2018-10-11 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis for the inhibitory effects of a novel reversible covalent ligand on PPAR gamma phosphorylation.
Sci Rep, 9, 2019
|
|
9QGF
 
 | | Crystal structure of an NADH-accepting ene reductase variant NostocER1-L1,5 mutant T354K | | Descriptor: | 1,2-ETHANEDIOL, 1-[(6~{R})-3,3-diethanoyl-6-methyl-cyclohexa-1,4-dien-1-yl]ethanone, All1865 protein, ... | | Authors: | Bischoff, D, Walla, B, Janowski, R, Maslakova, A, Niessing, D, Weuster-Botz, D. | | Deposit date: | 2025-03-13 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.444 Å) | | Cite: | Rational Introduction of Electrostatic Interactions at Crystal Contacts to Enhance Protein Crystallization of an Ene Reductase.
Biomolecules, 15, 2025
|
|
3C0A
 
 | | Lactobacillus CASEI Thymidylate Synthase Ternary Complex with DUMP and the Phtalimidic Derivative 14C in Multiple Binding Modes-Mode 2 | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-(2-chloropyridin-4-yl)-4-methyl-1H-isoindole-1,3(2H)-dione, Thymidylate synthase | | Authors: | Leone, R, Mangani, S, Cancian, L, Costi, M.P, Luciani, R, Ferrari, S. | | Deposit date: | 2008-01-19 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of the binding modes of N-phenylphthalimides inhibiting bacterial thymidylate synthase through X-ray crystallography screening
J.Med.Chem., 54, 2011
|
|
7LAZ
 
 | | Crystal structure of the first bromodomain (BD1) of human BRD3 bound to ERK5-IN-1 | | Descriptor: | 11-cyclopentyl-2-({2-ethoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 3 | | Authors: | Karim, M.R, Bikowitz, M.J, Schonbrunn, E. | | Deposit date: | 2021-01-07 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
3BYX
 
 | | Lactobacillus CASEI Thymidylate Synthase Ternary Complex with DUMP and the Phtalimidic Derivative C00 in Multiple Binding Modes | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-(4-hydroxybiphenyl-3-yl)-4-methyl-1H-isoindole-1,3(2H)-dione, Thymidylate synthase | | Authors: | Leone, R, Mangani, S, Cancian, L, Costi, M.P, Luciani, R, Ferrari, S. | | Deposit date: | 2008-01-16 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of the binding modes of N-phenylphthalimides inhibiting bacterial thymidylate synthase through X-ray crystallography screening
J.Med.Chem., 54, 2011
|
|
5G21
 
 | | Leishmania major N-myristoyltransferase in complex with a quinoline inhibitor (compound 26). | | Descriptor: | ETHYL 4-[(2-CYANOETHYL)SULFANYL]-6-{[6-(PIPERAZIN-1-YL), GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Goncalves, V, Brannigan, J.A, Laporte, A, Bell, A.S, Roberts, S.M, Wilkinson, A.J, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2016-04-06 | | Release date: | 2017-02-15 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided optimization of quinoline inhibitors of Plasmodium N-myristoyltransferase.
Medchemcomm, 8, 2017
|
|
4PSP
 
 | | Crystal Structure of GH29 family alpha-L-fucosidase from Fusarium graminearum in the open form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-fucosidase GH29, ... | | Authors: | Cao, H, Walton, J, Brumm, P, Phillips Jr, G.N. | | Deposit date: | 2014-03-07 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | Structure and Substrate Specificity of a Eukaryotic Fucosidase from Fusarium graminearum.
J.Biol.Chem., 289, 2014
|
|
7Q4C
 
 | | Local refinement structure of the C-domain of full-length, monomeric, soluble somatic angiotensin I-converting enzyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Lubbe, L, Sewell, B.T, Sturrock, E.D. | | Deposit date: | 2021-10-30 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Cryo-EM reveals mechanisms of angiotensin I-converting enzyme allostery and dimerization.
Embo J., 41, 2022
|
|
1XFI
 
 | | X-ray structure of gene product from Arabidopsis thaliana At2g17340 | | Descriptor: | MAGNESIUM ION, unknown protein | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-09-14 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure at 1.7 A resolution of the protein product of the At2g17340 gene from Arabidopsis thaliana.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
3BKN
 
 | | The structure of Mycobacterial bacterioferritin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bacterioferritin, MAGNESIUM ION, ... | | Authors: | Janowski, R, Auerbach-Nevo, T, Weiss, M.S. | | Deposit date: | 2007-12-07 | | Release date: | 2008-01-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Bacterioferritin from Mycobacterium smegmatis contains zinc in its di-nuclear site.
Protein Sci., 17, 2008
|
|
6I9Q
 
 | | Structure of the mouse CD98 heavy chain ectodomain | | Descriptor: | 1,2-ETHANEDIOL, 4F2 cell-surface antigen heavy chain, CHLORIDE ION | | Authors: | Schiefner, A, Deuschle, F.-C, Skerra, A. | | Deposit date: | 2018-11-24 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural differences between the ectodomains of murine and human CD98hc.
Proteins, 87, 2019
|
|
8GC1
 
 | |
9QNV
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-5-phenyl-[1,3]thiazolo[2,3-c][1,2,4]triazole, Bromodomain-containing protein 4, ... | | Authors: | Krojer, T, Martinez-Cartro, M, Picaud, S, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Barril, X, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-03-25 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.226 Å) | | Cite: | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound 2
To Be Published
|
|
