5JWA
 
 | | the structure of malaria PfNDH2 | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, FRAGMENT OF TRITON X-100, ... | | Authors: | Yu, Y, Yang, Y.Q, Li, X.L, Yu, J, Ge, J.P, Li, J, Rao, Y, Yang, M.J. | | Deposit date: | 2016-05-11 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Target Elucidation by Cocrystal Structures of NADH-Ubiquinone Oxidoreductase of Plasmodium falciparum (PfNDH2) with Small Molecule To Eliminate Drug-Resistant Malaria
J. Med. Chem., 60, 2017
|
|
3I63
 
 | | Peroxide Bound Toluene 4-Monooxygenase | | Descriptor: | FE (III) ION, HYDROGEN PEROXIDE, Toluene-4-monooxygenase system protein A, ... | | Authors: | Bailey, L.J, Fox, B.G. | | Deposit date: | 2009-07-06 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystallographic and catalytic studies of the peroxide-shunt reaction in a diiron hydroxylase.
Biochemistry, 48, 2009
|
|
9IWV
 
 | | Crystal structure of Lsd18 after incubation with the substrate | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Wang, Q, Kim, C.Y, Chen, X. | | Deposit date: | 2024-07-26 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanistic and molecular insights into iterative triepoxidation catalyzed by monooxygenase MonCI using a natural substrate analog.
Int.J.Biol.Macromol., 311, 2025
|
|
1APC
 
 | |
8YOQ
 
 | | Crystal structure of hFSP1 with both 6-OH-FAD and NADP (hFSP1-6-OH-FAD-NADP) | | Descriptor: | 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, Ferroptosis suppressor protein 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lan, H.Y, Gao, Y, Hong, T, Zhao, Z.Z, Chang, Z.H, Wang, Y.F, Wang, F. | | Deposit date: | 2024-03-13 | | Release date: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural insight into 6-OH-FAD-dependent activation of hFSP1 for ferroptosis suppression.
Cell Discov, 10, 2024
|
|
8YO8
 
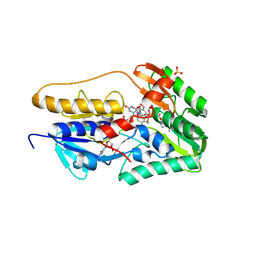 | | Crystal structure of hFSP1 with both 6-OH-FAD and NAD (hFSP1-6-OH-FAD-NAD) | | Descriptor: | 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, Ferroptosis suppressor protein 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lan, H.Y, Gao, Y, Hong, T, Zhao, Z.Z, Chang, Z.H, Wang, Y.F, Wang, F. | | Deposit date: | 2024-03-12 | | Release date: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into 6-OH-FAD-dependent activation of hFSP1 for ferroptosis suppression.
Cell Discov, 10, 2024
|
|
8YOX
 
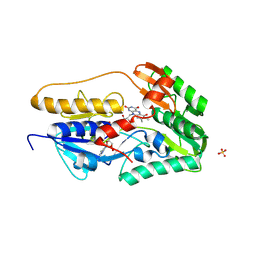 | | Crystal structure of hFSP1 with 6-OH-FAD (hFSP1-6-OH-FAD) | | Descriptor: | 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, Ferroptosis suppressor protein 1, SULFATE ION | | Authors: | Lan, H.Y, Gao, Y, Hong, T, Zhao, Z.Z, Chang, Z.H, Wang, Y.F, Wang, F. | | Deposit date: | 2024-03-14 | | Release date: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural insight into 6-OH-FAD-dependent activation of hFSP1 for ferroptosis suppression.
Cell Discov, 10, 2024
|
|
1NDL
 
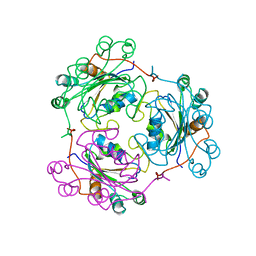 | | THE AWD NUCLEOTIDE DIPHOSPHATE KINASE FROM DROSOPHILA | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Janin, J, Chiadmi, M, Dumas, C, Lascu, I, Lebras, G, Morera, S, Veron, M. | | Deposit date: | 1993-11-27 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the Awd nucleotide diphosphate kinase from Drosophila.
Structure, 1, 1993
|
|
9CVU
 
 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain bound with 4-methyl-6-(3-((methylamino)methyl)-5-(trifluoromethyl)phenyl)pyridin-2-amine dihydrochloride | | Descriptor: | (6P)-4-methyl-6-{3-[(methylamino)methyl]-5-(trifluoromethyl)phenyl}pyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2024-07-29 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Truncated pyridinylbenzylamines: Potent, selective, and highly membrane permeable inhibitors of human neuronal nitric oxide synthase.
Bioorg.Med.Chem., 124, 2025
|
|
9CW1
 
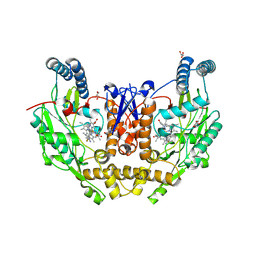 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain bound with 6-(2,3-difluoro-5-(2-(methylamino)ethyl)phenyl)-4-methylpyridin-2-amine dihydrochloride | | Descriptor: | (6M)-6-{2,3-difluoro-5-[2-(methylamino)ethyl]phenyl}-4-methylpyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2024-07-29 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Truncated pyridinylbenzylamines: Potent, selective, and highly membrane permeable inhibitors of human neuronal nitric oxide synthase.
Bioorg.Med.Chem., 124, 2025
|
|
1MMM
 
 | | DISTINCT METAL ENVIRONMENT IN IRON-SUBSTITUTED MANGANESE SUPEROXIDE DISMUTASE PROVIDES A STRUCTURAL BASIS OF METAL SPECIFICITY | | Descriptor: | FE (III) ION, HYDROXIDE ION, PROTEIN (IRON-SUBSTITUTED MANGANESE SUPEROXIDE DISMUTASE) | | Authors: | Edwards, R.A, Whittaker, M.M, Whittaker, J.W, Jameson, G.B, Baker, E.N. | | Deposit date: | 1998-08-26 | | Release date: | 1998-09-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Distinct Metal Environment in Fe-Substituted Manganese Superoxide Dismutase Provides a Structural Basis of Metal Specificity
J.Am.Chem.Soc., 120, 1998
|
|
5WP2
 
 | | 1.44 Angstrom crystal structure of CYP121 from Mycobacterium tuberculosis in complex with substrate and CN | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, CYANIDE ION, Mycocyclosin synthase, ... | | Authors: | Fielding, A, Dornevil, K, Liu, A. | | Deposit date: | 2017-08-03 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.439 Å) | | Cite: | Probing Ligand Exchange in the P450 Enzyme CYP121 from Mycobacterium tuberculosis: Dynamic Equilibrium of the Distal Heme Ligand as a Function of pH and Temperature.
J. Am. Chem. Soc., 139, 2017
|
|
6QO0
 
 | |
1SSO
 
 | | SOLUTION STRUCTURE AND DNA-BINDING PROPERTIES OF A THERMOSTABLE PROTEIN FROM THE ARCHAEON SULFOLOBUS SOLFATARICUS | | Descriptor: | SSO7D | | Authors: | Baumann, H, Knapp, S, Lundback, T, Ladenstein, R, Hard, T. | | Deposit date: | 1995-03-31 | | Release date: | 1995-05-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and DNA-binding properties of a thermostable protein from the archaeon Sulfolobus solfataricus.
Nat.Struct.Biol., 1, 1994
|
|
6ICP
 
 | | Pseudomonas putida CBB5 NdmA QL mutant with caffeine | | Descriptor: | CAFFEINE, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Kim, J.H, Kim, B.H, Kang, S.Y, Song, H.K. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Mechanistic Insights into Caffeine Degradation by the Bacterial N-Demethylase Complex.
J.Mol.Biol., 431, 2019
|
|
1CCN
 
 | | DIRECT NOE REFINEMENT OF CRAMBIN FROM 2D NMR DATA USING A SLOW-COOLING ANNEALING PROTOCOL | | Descriptor: | CRAMBIN | | Authors: | Bonvin, A.M.J.J, Rullmann, J.A.C, Lamerichs, R.M.J.N, Boelens, R, Kaptein, R. | | Deposit date: | 1993-04-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Direct NOE refinement of biomolecular structures using 2D NMR data
J.Biomol.NMR, 1, 1991
|
|
5LY1
 
 | | JMJD2A/ KDM4A COMPLEXED WITH NI(II) AND Macrocyclic PEPTIDE Inhibitor CP2 (13-mer) | | Descriptor: | CHLORIDE ION, CP2, GLYCEROL, ... | | Authors: | King, O.N.F, Chowdhury, R, Kawamura, A, Schofield, C.J. | | Deposit date: | 2016-09-23 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Highly selective inhibition of histone demethylases by de novo macrocyclic peptides.
Nat Commun, 8, 2017
|
|
6ICN
 
 | | Pseudomonas putida CBB5 NdmA with caffeine | | Descriptor: | CAFFEINE, COBALT (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Kim, J.H, Kim, B.H, Kang, S.Y, Song, H.K. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Mechanistic Insights into Caffeine Degradation by the Bacterial N-Demethylase Complex.
J.Mol.Biol., 431, 2019
|
|
7KPZ
 
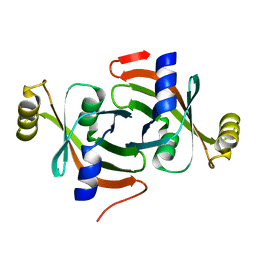 | |
7E0C
 
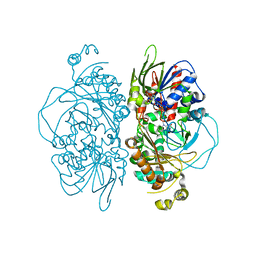 | | Structure of L-glutamate oxidase R305E mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-glutamate oxidase | | Authors: | Ito, N, Matsuo, S, Inagaki, K, Imada, K. | | Deposit date: | 2021-01-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A new l-arginine oxidase engineered from l-glutamate oxidase.
Protein Sci., 30, 2021
|
|
7E0D
 
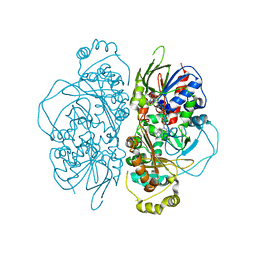 | | Structure of L-glutamate oxidase R305E mutant in complex with L-arginine | | Descriptor: | ARGININE, FLAVIN-ADENINE DINUCLEOTIDE, L-glutamate oxidase | | Authors: | Ito, N, Matsuo, S, Inagaki, K, Imada, K. | | Deposit date: | 2021-01-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A new l-arginine oxidase engineered from l-glutamate oxidase.
Protein Sci., 30, 2021
|
|
5IBO
 
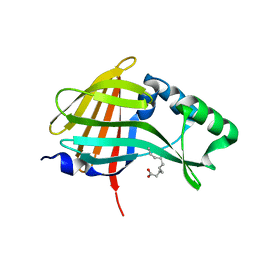 | | 1.95A resolution structure of NanoLuc luciferase | | Descriptor: | DECANOIC ACID, Oplophorus-luciferin 2-monooxygenase catalytic subunit | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Encell, L.P, Wood, K.V. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | To be determined
To be published
|
|
5ZMD
 
 | | Crystal structure of FTO in complex with m6dA modified ssDNA | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase FTO, DNA (5'-D(P*TP*CP*TP*(6MA)P*TP*AP*TP*CP*G)-3'), MANGANESE (II) ION, ... | | Authors: | Zhang, X, Wei, L.H, Luo, J, Xiao, Y, Liu, J, Zhang, W, Zhang, L, Jia, G.F. | | Deposit date: | 2018-04-02 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into FTO's catalytic mechanism for the demethylation of multiple RNA substrates.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
1BFY
 
 | | SOLUTION STRUCTURE OF REDUCED CLOSTRIDIUM PASTEURIANUM RUBREDOXIN, NMR, 20 STRUCTURES | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Bertini, I, Kurtz Junior, D.M, Eidsness, M.K, Liu, G, Luchinat, C, Rosato, A, Scott, R.A. | | Deposit date: | 1998-05-23 | | Release date: | 1999-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Reduced Clostridium Pasteurianum Rubredoxin
J.Biol.Inorg.Chem., 3, 1998
|
|
5JSK
 
 | | The 3D structure of [NiFeSe] hydrogenase from Desulfovibrio vulgaris Hildenborough in the reduced state at 0.95 Angstrom resolution | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE (II) ION, GLYCEROL, ... | | Authors: | Marques, M.C, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2016-05-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The direct role of selenocysteine in [NiFeSe] hydrogenase maturation and catalysis.
Nat. Chem. Biol., 13, 2017
|
|
