4M3C
 
 | | Structure of a binary complex between homologous tetrameric legume lectins from Butea monosperma and Spatholobus parviflorus seeds | | Descriptor: | CALCIUM ION, GAMMA-AMINO-BUTANOIC ACID, GLYCEROL, ... | | Authors: | Surya, S, Abhilash, J, Geethanandan, K, Sadasivan, C, Haridas, M. | | Deposit date: | 2013-08-06 | | Release date: | 2013-09-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a binary complex between homologous tetrameric legume lectins from Butea monosperma and Spatholobus parviflorus seeds
To be Published
|
|
5W4A
 
 | | C. japonica N-domain | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IMIDAZOLE, ... | | Authors: | Aoki, S.T, Bingman, C.A, Kimble, J. | | Deposit date: | 2017-06-09 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | C. elegans germ granules require both assembly and localized regulators for mRNA repression.
Nat Commun, 12, 2021
|
|
1B48
 
 | |
3FR4
 
 | | N-Benzyl-indolo carboxylic acids: Design and synthesis of potent and selective adipocyte Fatty-Acid Binding Protein (A-FABP) inhibitors | | Descriptor: | 9-[2-(trifluoromethyl)benzyl]-2,3,4,9-tetrahydro-1H-carbazole-8-carboxylic acid, Fatty acid-binding protein, adipocyte | | Authors: | Barf, T, Hammer, K, Lehmann, F, Haile, S, Axen, E, Medina, C, Uppenberg, J, Svensson, S, Rondahl, L, Lundb ck, T. | | Deposit date: | 2009-01-08 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | N-Benzyl-indolo carboxylic acids: Design and synthesis of potent and selective adipocyte fatty-acid binding protein (A-FABP) inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6NUJ
 
 | |
3T5E
 
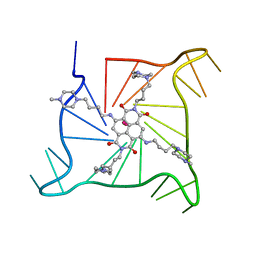 | | Crystal structure of an intramolecular human telomeric DNA G-quadruplex bound by the naphthalene diimide BMSG-SH-4 | | Descriptor: | 2,7-bis[4-(4-methylpiperazin-1-yl)butyl]-4,9-bis{[4-(4-methylpiperazin-1-yl)butyl]amino}benzo[lmn][3,8]phenanthroline-1 ,3,6,8(2H,7H)-tetrone, POTASSIUM ION, human telomeric DNA sequence | | Authors: | Collie, G.W, Promontorio, R, Parkinson, G.N. | | Deposit date: | 2011-07-27 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for telomeric g-quadruplex targeting by naphthalene diimide ligands.
J.Am.Chem.Soc., 134, 2012
|
|
5EJ8
 
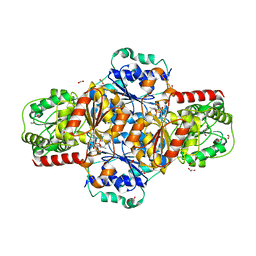 | | EcMenD-ThDP-Mn2+ complex structure soaked with 2-ketoglutarate for 2 min | | Descriptor: | (4S)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3lambda~5~-thiazol-2-yl}-4-hydroxybutanoic acid, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Song, H.G, Dong, C, Chen, Y.Z, Sun, Y.R, Guo, Z.H. | | Deposit date: | 2015-11-01 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | A Thiamine-Dependent Enzyme Utilizes an Active Tetrahedral Intermediate in Vitamin K Biosynthesis
J.Am.Chem.Soc., 138, 2016
|
|
4RX5
 
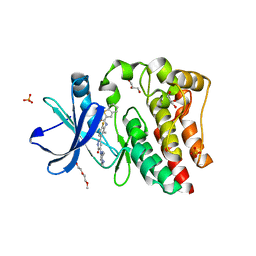 | | Bruton's tyrosine kinase (BTK) with pyridazinone compound 23 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, GLYCEROL, N-(6-fluoro-2-methyl-3-{5-[(5-methyl-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazin-2-yl)amino]-6-oxo-1,6-dihydropyridazin-3-yl}phenyl)-1-benzothiophene-2-carboxamide, ... | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2014-12-08 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.356 Å) | | Cite: | Discovery of highly potent and selective Bruton's tyrosine kinase inhibitors: Pyridazinone analogs with improved metabolic stability.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
3FCK
 
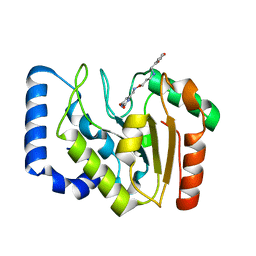 | | Complex of UNG2 and a fragment-based design inhibitor | | Descriptor: | 3-({[3-({[(1E)-(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)methylidene]amino}oxy)propyl]amino}methyl)benzoic acid, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Chung, S, Parker, J.B, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2008-11-21 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Impact of linker strain and flexibility in the design of a fragment-based inhibitor
Nat.Chem.Biol., 5, 2009
|
|
4NJS
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with non-peptidic inhibitor, GRL008 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-carbamoylphenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Garimella, H, Kaufman, J.D, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
3T95
 
 | | Crystal structure of LsrB from Yersinia pestis complexed with autoinducer-2 | | Descriptor: | (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran, Autoinducer 2-binding protein lsrB | | Authors: | Kavanaugh, J.S, Gakhar, L, Horswill, A.R. | | Deposit date: | 2011-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of LsrB from Yersinia pestis complexed with autoinducer-2.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
4BVX
 
 | | Crystal structure of the AIMP3-MRS N-terminal domain complex with I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, EUKARYOTIC TRANSLATION ELONGATION FACTOR 1 EPSILON-1, METHIONINE--TRNA LIGASE, ... | | Authors: | Cho, H.Y, Seo, W.W, Cho, H.J, Kang, B.S. | | Deposit date: | 2013-06-29 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Assembly of Multi-tRNA Synthetase Complex Via Heterotetrameric Glutathione Transferase-Homology Domains.
J.Biol.Chem., 290, 2015
|
|
3ALU
 
 | | Crystal structure of CEL-IV complexed with Raffinose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Lectin CEL-IV, ... | | Authors: | Hatakeyama, T, Hozawa, T, Ishii, K, Kamiya, T, Goda, S, Kusunoki, M, Unno, H. | | Deposit date: | 2010-08-07 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Galactose recognition by a tetrameric C-type lectin, CEL-IV, containing the EPN carbohydrate recognition motif
J.Biol.Chem., 286, 2011
|
|
5V8O
 
 | | Discovery of a high affinity inhibitor of cGAS | | Descriptor: | 5-phenyltetrazolo[1,5-a]pyrimidin-7-ol, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Hall, J. | | Deposit date: | 2017-03-22 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of PF-06928215 as a high affinity inhibitor of cGAS enabled by a novel fluorescence polarization assay.
PLoS ONE, 12, 2017
|
|
3ALT
 
 | | Crystal structure of CEL-IV complexed with Melibiose | | Descriptor: | CALCIUM ION, Lectin CEL-IV, C-type, ... | | Authors: | Hatakeyama, T, Hozawa, T, Ishii, K, Kamiya, T, Goda, S, Kusunoki, M, Unno, H. | | Deposit date: | 2010-08-07 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Galactose recognition by a tetrameric C-type lectin, CEL-IV, containing the EPN carbohydrate recognition motif
J.Biol.Chem., 286, 2011
|
|
3ALS
 
 | | Crystal structure of CEL-IV | | Descriptor: | CALCIUM ION, Lectin CEL-IV, C-type | | Authors: | Hatakeyama, T, Hozawa, T, Ishii, K, Kamiya, T, Goda, S, Kusunoki, M, Unno, H. | | Deposit date: | 2010-08-07 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Galactose recognition by a tetrameric C-type lectin, CEL-IV, containing the EPN carbohydrate recognition motif
J.Biol.Chem., 286, 2011
|
|
4NJT
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Yedidi, R.S, Garimella, H, Chang, S.B, Kaufman, J.D, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
2R0H
 
 | | Fungal lectin CGL3 in complex with chitotriose (chitotetraose) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CGL3 lectin | | Authors: | Waelti, M.A, Walser, P.J, Thore, S, Gruenler, A, Ban, N, Kuenzler, M, Aebi, M. | | Deposit date: | 2007-08-20 | | Release date: | 2008-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Chitotetraose Coordination by CGL3, a Novel Galectin-Related Protein from Coprinopsis cinerea
J.Mol.Biol., 379, 2008
|
|
4NJU
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with tipranavir | | Descriptor: | N-(3-{(1R)-1-[(6R)-4-HYDROXY-2-OXO-6-PHENETHYL-6-PROPYL-5,6-DIHYDRO-2H-PYRAN-3-YL]PROPYL}PHENYL)-5-(TRIFLUOROMETHYL)-2-PYRIDINESULFONAMIDE, Protease | | Authors: | Yedidi, R.S, Garimella, H, Kaufman, J.D, Das, D, Wingfield, P.T, Mitsuya, H. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
4NJV
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with ritonavir | | Descriptor: | Protease, RITONAVIR | | Authors: | Yedidi, R.S, Garimella, H, Chang, S.B, Kaufman, J.D, Das, D, Wingfield, P.T, Mitsuya, H. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
3PGT
 
 | | CRYSTAL STRUCTURE OF HGSTP1-1[I104] COMPLEXED WITH THE GSH CONJUGATE OF (+)-ANTI-BPDE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-4-[1-(CARBOXYMETHYL-CARBAMOYL)-2-(9-HYDROXY-7,8-DIOXO-7,8,9,10-TETRAHYDRO-BENZO[DEF]CHRYSEN-10-YLSULFANYL)-ETHYLCARBAMOYL]-BUTYRIC ACID, PROTEIN (GLUTATHIONE S-TRANSFERASE), ... | | Authors: | Ji, X, Xiao, B. | | Deposit date: | 1999-03-22 | | Release date: | 1999-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure and function of residue 104 and water molecules in the xenobiotic substrate-binding site in human glutathione S-transferase P1-1.
Biochemistry, 38, 1999
|
|
5DLA
 
 | | Structure of Tetragonal Lysozyme solved by UWO Students | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Bednarski, R, Cirricione, N, Greco, A, Hodgson, R, Kent, S, McGowan, J, Notherm, B, Patt, M, Vue, L, Bianchetti, C.M. | | Deposit date: | 2015-09-04 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structure of Tetragonal Lysozyme in complex with Iodine solved by UWO Students
To Be Published
|
|
4X12
 
 | | JC Polyomavirus genotype 3 VP1 in complex with GD1b oligosaccharide | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Major capsid protein, ... | | Authors: | Stroh, L.J, Stehle, T. | | Deposit date: | 2014-11-24 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Greater Affinity of JC Polyomavirus Capsid for alpha 2,6-Linked Lactoseries Tetrasaccharide c than for Other Sialylated Glycans Is a Major Determinant of Infectivity.
J.Virol., 89, 2015
|
|
4X13
 
 | | JC Polyomavirus genotype 3 VP1 in complex with LSTc pentasaccharide | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Major capsid protein, ... | | Authors: | Stroh, L.J, Stehle, T. | | Deposit date: | 2014-11-24 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Greater Affinity of JC Polyomavirus Capsid for alpha 2,6-Linked Lactoseries Tetrasaccharide c than for Other Sialylated Glycans Is a Major Determinant of Infectivity.
J.Virol., 89, 2015
|
|
2ZK1
 
 | | Human peroxisome proliferator-activated receptor gamma ligand binding domain complexed with 15-deoxy-delta12,14-prostaglandin J2 | | Descriptor: | (5E,14E)-11-oxoprosta-5,9,12,14-tetraen-1-oic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Waku, T, Shiraki, T, Oyama, T, Fujimoto, Y, Morikawa, K. | | Deposit date: | 2008-03-12 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural insight into PPARgamma activation through covalent modification with endogenous fatty acids
J.Mol.Biol., 385, 2009
|
|
