2XC1
 
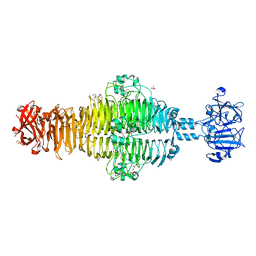 | | Full-length Tailspike Protein Mutant Y108W of Bacteriophage P22 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, BIFUNCTIONAL TAIL PROTEIN, CALCIUM ION, ... | | Authors: | Mueller, J.J, Seul, A, Seckler, R, Heinemann, U. | | Deposit date: | 2010-04-15 | | Release date: | 2011-05-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Bacteriophage P22 Tailspike: Structure of the Complete Protein and Function of the Interdomain Linker
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8G0D
 
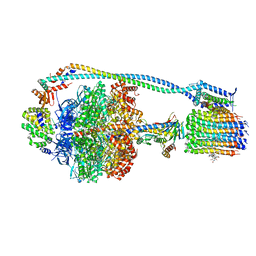 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase rotational state 2 (backbone model) | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
4Z6D
 
 | |
8TRG
 
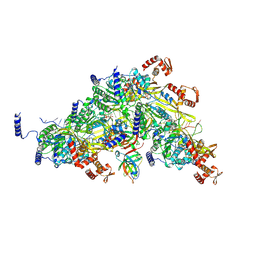 | | Structure of full-length LexA bound to a RecA filament | | Descriptor: | DNA (27-MER), LexA repressor, MAGNESIUM ION, ... | | Authors: | Cory, M.B, Li, A, Kohli, R.M. | | Deposit date: | 2023-08-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | The LexA-RecA* structure reveals a cryptic lock-and-key mechanism for SOS activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4Z6L
 
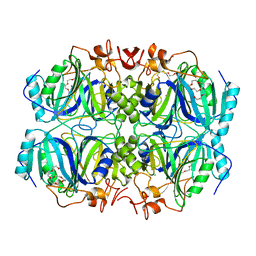 | | Structure of H200E variant of Homoprotocatechuate 2,3-Dioxygenase from B.fuscum at 1.65 Ang resolution | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (II) ION, ... | | Authors: | Kovaleva, E.G, Lipscomb, J.D. | | Deposit date: | 2015-04-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for Substrate and Oxygen Activation in Homoprotocatechuate 2,3-Dioxygenase: Roles of Conserved Active Site Histidine 200.
Biochemistry, 54, 2015
|
|
6WP1
 
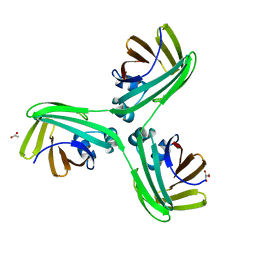 | |
4Z6Z
 
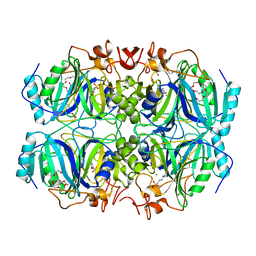 | | Structure of Homoprotocatechuate 2,3-Dioxygenase from B.fuscum in complex with 4-sulfonyl catechol at 1.52 Ang resolution | | Descriptor: | 3,4-dihydroxybenzenesulfonic acid, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CALCIUM ION, ... | | Authors: | Kovaleva, E.G, Lipscomb, J.D. | | Deposit date: | 2015-04-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural Basis for Substrate and Oxygen Activation in Homoprotocatechuate 2,3-Dioxygenase: Roles of Conserved Active Site Histidine 200.
Biochemistry, 54, 2015
|
|
1NLF
 
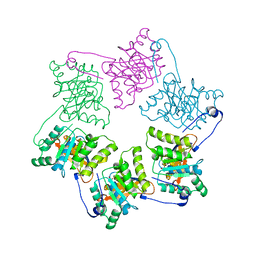 | | Crystal Structure of DNA Helicase RepA in complex with sulfate at 1.95 A resolution | | Descriptor: | Regulatory protein repA, SULFATE ION | | Authors: | Xu, H, Strater, N, Schroeder, W, Bottcher, C, Ludwig, K, Saenger, W. | | Deposit date: | 2003-01-07 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of DNA helicase RepA in complex with sulfate at 1.95 A resolution implicates structural changes to an "open" form.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4OH1
 
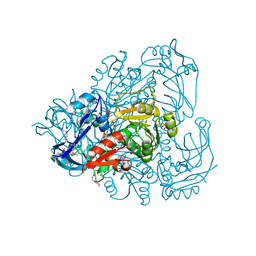 | |
4GX2
 
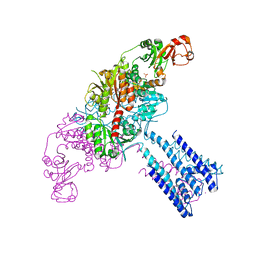 | | GsuK channel bound to NAD | | Descriptor: | CALCIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Kong, C, Zeng, W, Ye, S, Chen, L, Sauer, D.B, Lam, Y, Derebe, M.G, Jiang, Y. | | Deposit date: | 2012-09-03 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Distinct gating mechanisms revealed by the structures of a multi-ligand gated K(+) channel.
elife, 1, 2012
|
|
4GZA
 
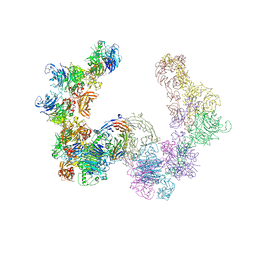 | | Complex of mouse Plexin A2 - Semaphorin 3A - Neuropilin-1 | | Descriptor: | CALCIUM ION, Neuropilin-1, Plexin-A2, ... | | Authors: | Janssen, B.J.C, Malinauskas, T, Siebold, C, Jones, E.Y. | | Deposit date: | 2012-09-06 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Neuropilins lock secreted semaphorins onto plexins in a ternary signaling complex.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2Y1R
 
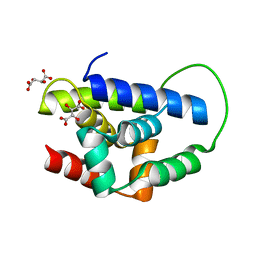 | | Structure of MecA121 & ClpC N-domain complex | | Descriptor: | ADAPTER PROTEIN MECA 1, NEGATIVE REGULATOR OF GENETIC COMPETENCE CLPC/MECB, S,R MESO-TARTARIC ACID | | Authors: | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Structure and Mechanism of the Hexameric Meca-Clpc Molecular Machine.
Nature, 471, 2011
|
|
8G46
 
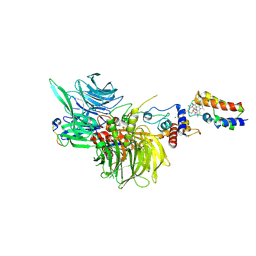 | | Cryo-EM structure of DDB1deltaB-DDA1-DCAF16-BRD4(BD2)-MMH2 | | Descriptor: | Bromodomain-containing protein 4, DDB1- and CUL4-associated factor 16, DET1- and DDB1-associated protein 1, ... | | Authors: | Ma, M.W, Hunkeler, M, Jin, C.Y, Fischer, E.S. | | Deposit date: | 2023-02-08 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Template-assisted covalent modification of DCAF16 underlies activity of BRD4 molecular glue degraders.
Biorxiv, 2023
|
|
6WPA
 
 | |
4GYN
 
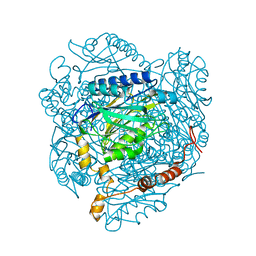 | | The E142L mutant of the amidase from Geobacillus pallidus | | Descriptor: | Aliphatic amidase, CHLORIDE ION | | Authors: | Weber, B.W, Sewell, B.T, Kimani, S.W, Varsani, A, Cowan, D.A, Hunter, R. | | Deposit date: | 2012-09-05 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The mechanism of the amidases: mutating the glutamate adjacent to the catalytic triad inactivates the enzyme due to substrate mispositioning.
J.Biol.Chem., 288, 2013
|
|
5NVU
 
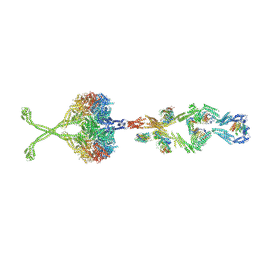 | | Full length human cytoplasmic dynein-1 in the phi-particle conformation | | Descriptor: | Dynein intermediate chain, Dynein light intermediate chain, Dynein motor domain, ... | | Authors: | Zhang, K, Foster, H.E, Carter, A.P. | | Deposit date: | 2017-05-04 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Cryo-EM Reveals How Human Cytoplasmic Dynein Is Auto-inhibited and Activated.
Cell, 169, 2017
|
|
8TNS
 
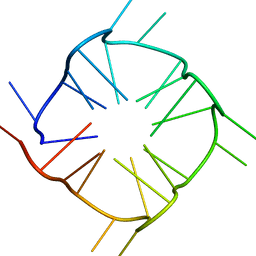 | |
2XXW
 
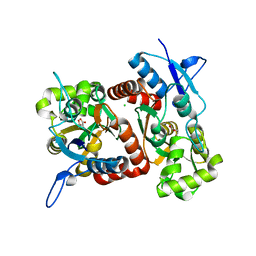 | | Crystal structure of the GluK2 (GluR6) D776K LBD dimer in complex with glutamate | | Descriptor: | CHLORIDE ION, GLUTAMATE RECEPTOR, IONOTROPIC KAINATE 2, ... | | Authors: | Nayeem, N, Mayans, O, Green, T. | | Deposit date: | 2010-11-12 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational Flexibility of the Ligand-Binding Domain Dimer in Kainate Receptor Gating and Desensitization
J.Neurosci., 31, 2011
|
|
6WDW
 
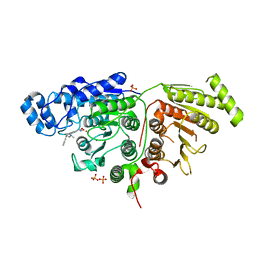 | |
3W3G
 
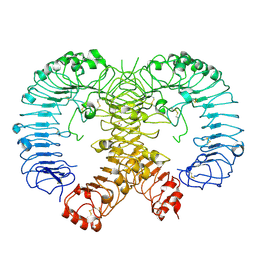 | | Crystal structure of human TLR8 (unliganded form) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2012-12-21 | | Release date: | 2013-04-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural reorganization of the Toll-like receptor 8 dimer induced by agonistic ligands
Science, 339, 2013
|
|
5OA4
 
 | | Fe(II)/(alpha)ketoglutarate-dependent dioxygenase AsqJ_V72I mutant in complex with 4-methoxycyclopeptin (1) | | Descriptor: | (3S)-3-(4-methoxybenzyl)-4-methyl-3,4-dihydro-1H-1,4-benzodiazepine-2,5-dione, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Groll, M, Braeuer, A, Kaila, V.R.I. | | Deposit date: | 2017-06-20 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Catalytic mechanism and molecular engineering of quinolone biosynthesis in dioxygenase AsqJ.
Nat Commun, 9, 2018
|
|
4Z9V
 
 | | TCTP contains a BH3-like domain, which instead of inhibiting, activates Bcl-xL | | Descriptor: | BICARBONATE ION, Bcl-2-like protein 1,APOPTOSIS REGULATOR BCL-XL, SODIUM ION, ... | | Authors: | Cura, V, Agez, M, Thebault, S, Cavarelli, J. | | Deposit date: | 2015-04-12 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | TCTP contains a BH3-like domain, which instead of inhibiting, activates Bcl-xL.
Sci Rep, 6, 2016
|
|
8GCA
 
 | | Mouse acidic mammalian chitinase, catalytic domain in complex with N,N',N''-triacetylchitotriose at pH 4.74 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acidic mammalian chitinase, ... | | Authors: | Diaz, R.E, Fraser, J.S. | | Deposit date: | 2023-03-01 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of ligand binding and pH-specific enzymatic activity of mouse Acidic Mammalian Chitinase.
Biorxiv, 2024
|
|
2Y5L
 
 | | orally active aminopyridines as inhibitors of tetrameric fructose 1,6- bisphosphatase | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATASE 1, N-{[(2Z)-5-bromo-1,3-thiazol-2(3H)-ylidene]carbamoyl}-3-chlorobenzenesulfonamide | | Authors: | ruf, a, hebeisen, p, haap, w, kuhn, b, mohr, p, wessel, h.p, zutter, u, kirchner, s, benz, j, joseph, c, alvarez-sanchez, r, gubler, m, schott, b, benardeau, a, tozzo, e, kitas, e. | | Deposit date: | 2011-01-14 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Orally Active Aminopyridines as Inhibitors of Tetrameric Fructose-1,6-Bisphosphatase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2Y1M
 
 | | Structure of native c-Cbl | | Descriptor: | CALCIUM ION, E3 UBIQUITIN-PROTEIN LIGASE, ZINC ION | | Authors: | Dou, H, Sibbet, G.J, Huang, D.T. | | Deposit date: | 2010-12-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural Basis for Autoinhibition and Phosphorylation-Dependent Activation of C-Cbl.
Nat.Struct.Mol.Biol., 19, 2012
|
|
