8TPB
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)-2-chloroacetamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
7OVD
 
 | | Human soluble adenylyl cyclase in complex with the inhibitor TDI10229 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-6-[1,5-dimethyl-4-(phenylmethyl)pyrazol-3-yl]pyrimidin-2-amine, ACETATE ION, ... | | Authors: | Steegborn, C, Quast, J. | | Deposit date: | 2021-06-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of TDI-10229: A Potent and Orally Bioavailable Inhibitor of Soluble Adenylyl Cyclase (sAC, ADCY10).
Acs Med.Chem.Lett., 12, 2021
|
|
8TPH
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | (3R)-N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-3-hydroxybutanamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8TPI
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-2-hydroxy-2-methylpropanamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
7OMN
 
 | | Anti-EphA1 JD1-1 VH domain | | Descriptor: | GLYCEROL, IMIDAZOLE, JD1-1 VH domain | | Authors: | Ereno Orbea, J, Nilvebrant, J, Sidhu, S, Julien, J.P. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Systematic Engineering of Optimized Autonomous Heavy-Chain Variable Domains.
J.Mol.Biol., 433, 2021
|
|
8FQJ
 
 | | LSD1-CoREST in complex with T14, short soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2S,3R,4R)-2,3,4-trihydroxy-5-[(1R,3S,3aS,13R)-1-hydroxy-10,11-dimethyl-3-{4-[(5-methyl-1,3,4-thiadiazol-2-yl)carbamoyl]phenyl}-4,6-dioxo-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]pentyl dihydrogen diphosphate | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2023-01-06 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
To be published
|
|
6FKZ
 
 | |
7P46
 
 | | Crystal Structure of Xanthomonas campestris Tryptophan 2,3-dioxygenase (TDO) | | Descriptor: | (2S)-2-amino-4-(2-aminophenyl)-4-oxobutanoic acid, CYANIDE ION, GLYCEROL, ... | | Authors: | Kwon, H, Basran, J, Booth, E.S, Campbell, L.P, Thackray, S.J, Moody, P.C.E, Mowat, C.G, Raven, E.L. | | Deposit date: | 2021-07-09 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of l-kynurenine to X. campestris tryptophan 2,3-dioxygenase.
J.Inorg.Biochem., 225, 2021
|
|
8FDV
 
 | | LSD1-CoREST in complex N-formyl FAD and SNAG peptide | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, Zinc finger protein SNAI1, ... | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2022-12-05 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
to be published
|
|
8TPD
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | 3C-like proteinase nsp5, N-[(1R)-2-(benzylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-[3-(2-chloroacetamido)phenyl]furan-2-carboxamide | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8FJ6
 
 | | LSD1-CoREST in complex with T108, long soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4S)-5-[7,8-dimethyl-2,4-dioxo-5-{3-[3-(phenylcarbamoyl)phenyl]propanoyl}-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2022-12-19 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
to be published
|
|
8TPF
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-3-hydroxypropanamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
7P9T
 
 | | Crystal structure of CD73 in complex with dCMP in the open form | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 5'-nucleotidase, CALCIUM ION, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-27 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
4M6U
 
 | |
8G5W
 
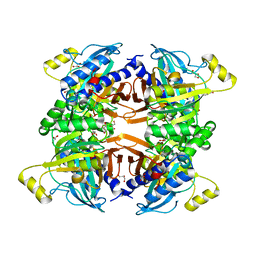 | | Structure of the Class II Fructose-1,6-Bisphophatase from Francisella tularensis complexed with native metal cofactor Mn++ | | Descriptor: | Fructose-1,6-bisphosphatase, GLYCEROL, MANGANESE (II) ION | | Authors: | Abad-Zapatero, C, Selezneva, A.I. | | Deposit date: | 2023-02-14 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New structures of Class II Fructose-1,6-Bisphosphatase from Francisella tularensis provide a framework for a novel catalytic mechanism for the entire class.
Plos One, 18, 2023
|
|
7PB5
 
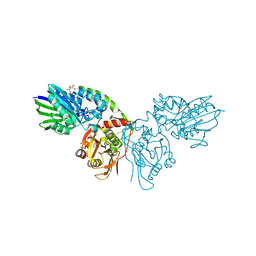 | | Crystal structure of CD73 in complex with UMP in the open form | | Descriptor: | 5'-nucleotidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-30 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
7PBA
 
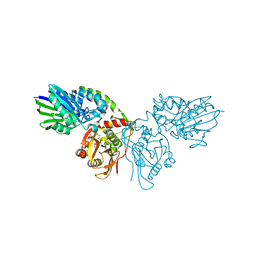 | | Crystal structure of CD73 in complex with IMP in the open form | | Descriptor: | 5'-nucleotidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-08-01 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
8T3S
 
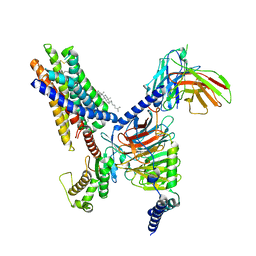 | | Cryo-EM structure of the Butyrate bound FFA2-Gq complex | | Descriptor: | CHOLESTEROL, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
4LNI
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of the transition state complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutamine synthetase, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, ... | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | Deposit date: | 2013-07-11 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5793 Å) | | Cite: | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
6FE1
 
 | |
7PA4
 
 | | Crystal structure of CD73 in complex with CMP in the open form | | Descriptor: | 5'-nucleotidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-28 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
8TPC
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | N-[(1R)-2-(benzylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-[4-(2-chloroacetamido)phenyl]furan-2-carboxamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
7OWY
 
 | | Crystal structure of human mitochondrial ferritin (hMTF) Fe(II)-loaded for 3 minutes showing a peroxide anion as bridging species of iron ions in the ferroxidase site | | Descriptor: | CHLORIDE ION, FE (II) ION, Ferritin, ... | | Authors: | Pozzi, C, Ciambellotti, S, Tassone, G, Turano, P, Mangani, S. | | Deposit date: | 2021-06-21 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Iron Binding in the Ferroxidase Site of Human Mitochondrial Ferritin.
Chemistry, 27, 2021
|
|
4LKO
 
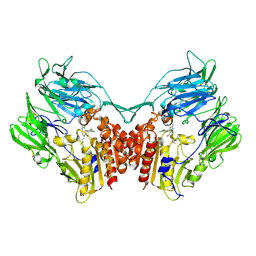 | | Crystal structure of human DPP-IV in complex with BMS-744891 | | Descriptor: | 3-(aminomethyl)-4-(2,4-dichlorophenyl)-6-(2-methoxyethyl)-2-methyl-6,7-dihydro-5H-pyrrolo[3,4-b]pyridin-5-one, Dipeptidyl peptidase 4 | | Authors: | Klei, H.E. | | Deposit date: | 2013-07-08 | | Release date: | 2013-09-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Optimization of Activity, Selectivity, and Liability Profiles in 5-Oxopyrrolopyridine DPP4 Inhibitors Leading to Clinical Candidate (Sa)-2-(3-(Aminomethyl)-4-(2,4-dichlorophenyl)-2-methyl-5-oxo-5H-pyrrolo[3,4-b]pyridin-6(7H)-yl)-N,N-dimethylacetamide (BMS-767778).
J.Med.Chem., 56, 2013
|
|
7PBB
 
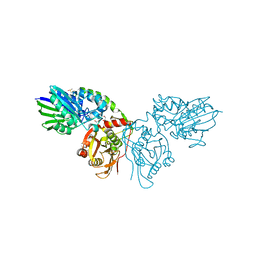 | |
