2FCL
 
 | |
5CJX
 
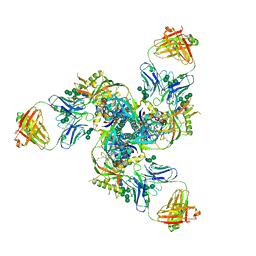 | |
7JHK
 
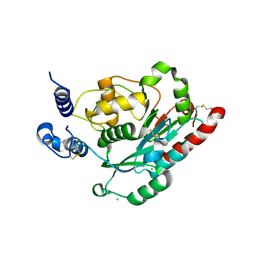 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 in unliganded form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hao, Y, Huang, X. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3436 Å) | | Cite: | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
4JNK
 
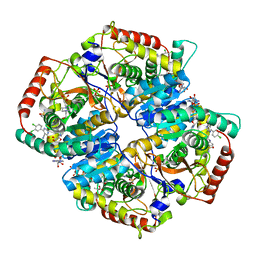 | | Lactate Dehydrogenase A in complex with inhibitor compound 22 | | Descriptor: | (2R)-2-{[5-cyano-4-(3,4-dichlorophenyl)-6-oxo-1,6-dihydropyrimidin-2-yl]sulfanyl}-N-(4-sulfamoylphenyl)propanamide, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2013-03-15 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Identification of substituted 2-thio-6-oxo-1,6-dihydropyrimidines as inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5TIQ
 
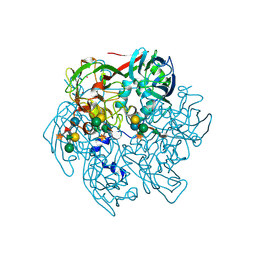 | | The Structure of the Major Capsid protein of PBCV-1 | | Descriptor: | 6-deoxy-2,3-di-O-methyl-alpha-L-mannopyranose-(1-2)-beta-L-rhamnopyranose-(1-4)-beta-D-xylopyranose-(1-4)-[alpha-D-mannopyranose-(1-3)-alpha-D-rhamnopyranose-(1-3)][alpha-D-galactopyranose-(1-2)]alpha-L-fucopyranose-(1-3)-[beta-D-xylopyranose-(1-4)]beta-D-glucopyranose, MERCURY (II) ION, Major capsid protein, ... | | Authors: | Klose, T, De Castro, C, Speciale, I, Molinaro, A, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2016-10-03 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.537 Å) | | Cite: | Structure of the chlorovirus PBCV-1 major capsid glycoprotein determined by combining crystallographic and carbohydrate molecular modeling approaches.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3N8X
 
 | | Crystal Structure of Cyclooxygenase-1 in Complex with Nimesulide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-NITRO-2-PHENOXYMETHANESULFONANILIDE, ... | | Authors: | Lee, J.Y. | | Deposit date: | 2010-05-28 | | Release date: | 2010-07-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Comparison of Cyclooxygenase-1 Crystal Structures: Cross-Talk between Monomers Comprising Cyclooxygenase-1 Homodimers
Biochemistry, 49, 2010
|
|
2F9K
 
 | | Crystal structure of human FPPS in complex with Zoledronate and Zn2+ | | Descriptor: | Farnesyl Diphosphate Synthase, PHOSPHATE ION, ZINC ION, ... | | Authors: | Rondeau, J.-M, Bitsch, F, Bourgier, E, Geiser, M, Hemmig, R, Kroemer, M, Lehmann, S, Ramage, P, Rieffel, S, Strauss, A, Green, J.R, Jahnke, W. | | Deposit date: | 2005-12-06 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for the exceptional in vivo efficacy of bisphosphonate drugs.
Chemmedchem, 1, 2006
|
|
2AYH
 
 | | CRYSTAL AND MOLECULAR STRUCTURE AT 1.6 ANGSTROMS RESOLUTION OF THE HYBRID BACILLUS ENDO-1,3-1,4-BETA-D-GLUCAN 4-GLUCANOHYDROLASE H(A16-M) | | Descriptor: | 1,3-1,4-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, CALCIUM ION | | Authors: | Hahn, M, Keitel, T, Heinemann, U. | | Deposit date: | 1995-02-02 | | Release date: | 1995-03-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal and molecular structure at 0.16-nm resolution of the hybrid Bacillus endo-1,3-1,4-beta-D-glucan 4-glucanohydrolase H(A16-M).
Eur.J.Biochem., 232, 1995
|
|
2GPI
 
 | |
3BZU
 
 | | Crystal structure of human 11-beta-hydroxysteroid dehydrogenase(HSD1) in complex with NADP and thiazolone inhibitor | | Descriptor: | (5S)-2-{[(1S)-1-(2-fluorophenyl)ethyl]amino}-5-methyl-5-(trifluoromethyl)-1,3-thiazol-4(5H)-one, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Min, X, Sudom, A, Xu, H, Wang, Z, Walker, N.P. | | Deposit date: | 2008-01-18 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural characterization and pharmacodynamic effects of an orally active 11beta-hydroxysteroid dehydrogenase type 1 inhibitor.
Chem.Biol.Drug Des., 71, 2008
|
|
3D4N
 
 | | Crystal Structure of Human 11-beta-Hydroxysteroid Dehydrogenase (HSD1) in Complex with Sulfonamide Inhibitor | | Descriptor: | 1-{[(3R)-3-methyl-4-({4-[(1S)-2,2,2-trifluoro-1-hydroxy-1-methylethyl]phenyl}sulfonyl)piperazin-1-yl]methyl}cyclopropanecarboxamide, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, Z, Liu, J, Sudom, A, Walker, N.P. | | Deposit date: | 2008-05-14 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Novel, Potent Benzamide Inhibitors of 11beta-Hydroxysteroid Dehydrogenase Type 1 (11beta-HSD1) Exhibiting Oral Activity in an Enzyme Inhibition ex Vivo Model
J.Med.Chem., 51, 2008
|
|
2YIO
 
 | |
4HMN
 
 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 5 in complex with (4-(4-Chlorophenyl)piperazin-1-yl)(morpholino)methanone (24) | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Turnbull, A.P, Flanagan, J.U, Atwell, G.J, Heinrich, D.M, Jamieson, S.M.F, Brooke, D.G, Silva, S, Rigoreau, L.J.M, Trivier, E, Soudy, C, Samlal, S.S, Owen, P.J, Schroeder, E, Raynham, T, Denny, W.A. | | Deposit date: | 2012-10-18 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Morpholylureas are a new class of potent and selective inhibitors of the type 5 17-beta-hydroxysteroid dehydrogenase (AKR1C3).
Bioorg.Med.Chem., 22, 2014
|
|
5TIP
 
 | | The Structure of the Major Capsid protein of PBCV-1 | | Descriptor: | 6-deoxy-2,3-di-O-methyl-alpha-L-mannopyranose-(1-2)-beta-L-rhamnopyranose-(1-4)-beta-D-xylopyranose-(1-4)-[alpha-D-mannopyranose-(1-3)-alpha-D-rhamnopyranose-(1-3)][alpha-D-galactopyranose-(1-2)]alpha-L-fucopyranose-(1-3)-[beta-D-xylopyranose-(1-4)]beta-D-glucopyranose, MERCURY (II) ION, Major capsid protein, ... | | Authors: | Klose, T, De Castro, C, Speciale, I, Molinaro, A, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2016-10-03 | | Release date: | 2017-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the chlorovirus PBCV-1 major capsid glycoprotein determined by combining crystallographic and carbohydrate molecular modeling approaches.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1AOE
 
 | | CANDIDA ALBICANS DIHYDROFOLATE REDUCTASE COMPLEXED WITH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE (NADPH) AND 1,3-DIAMINO-7-(1-ETHYEPROPYE)-7H-PYRRALO-[3,2-F]QUINAZOLINE (GW345) | | Descriptor: | 7-(1-ETHYL-PROPYL)-7H-PYRROLO-[3,2-F]QUINAZOLINE-1,3-DIAMINE, DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Whitlow, M, Howard, A.J, Stewart, D. | | Deposit date: | 1997-07-02 | | Release date: | 1998-01-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic studies of Candida albicans dihydrofolate reductase. High resolution structures of the holoenzyme and an inhibited ternary complex.
J.Biol.Chem., 272, 1997
|
|
2EAE
 
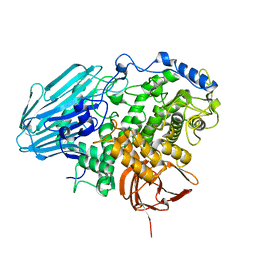 | | Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complexes with products | | Descriptor: | Alpha-fucosidase, CALCIUM ION, alpha-L-fucopyranose, ... | | Authors: | Nagae, M, Tsuchiya, A, Katayama, T, Yamamoto, K, Wakatsuki, S, Kato, R. | | Deposit date: | 2007-01-31 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis on the catalytic reaction mechanism of novel 1,2-alpha-L-fucosidase (AFCA) from Bifidobacterium bifidum
J.Biol.Chem., 282, 2007
|
|
5U7O
 
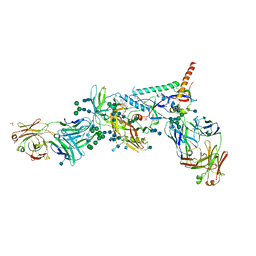 | | Crystal Structure of HIV-1 BG505 SOSIP.664 Prefusion Env Trimer Bound to Small Molecule HIV-1 Entry Inhibitor BMS-626529 in Complex with Human Antibodies PGT122 and 35O22 at 3.8 Angstrom | | Descriptor: | 1-[4-(benzenecarbonyl)piperazin-1-yl]-2-[4-methoxy-7-(3-methyl-1H-1,2,4-triazol-1-yl)-1H-pyrrolo[2,3-c]pyridin-3-yl]ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pancera, M, Lai, Y.-T, Kwong, P.D. | | Deposit date: | 2016-12-12 | | Release date: | 2017-08-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.031 Å) | | Cite: | Crystal structures of trimeric HIV envelope with entry inhibitors BMS-378806 and BMS-626529.
Nat. Chem. Biol., 13, 2017
|
|
2FJS
 
 | |
2F8C
 
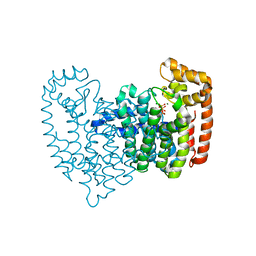 | | Crystal structure of FPPS in complex with Zoledronate | | Descriptor: | Farnesyl Diphosphate Synthase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Rondeau, J.-M, Bitsch, F, Bourgier, E, Geiser, M, Hemmig, R, Kroemer, M, Lehmann, S, Ramage, P, Rieffel, S, Strauss, A, Green, J.R, Jahnke, W. | | Deposit date: | 2005-12-02 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the exceptional in vivo efficacy of bisphosphonate drugs.
Chemmedchem, 1, 2006
|
|
2F8Z
 
 | | Crystal structure of human FPPS in complex with zoledronate and isopentenyl diphosphate | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Farnesyl Diphosphate Synthase, MAGNESIUM ION, ... | | Authors: | Rondeau, J.-M, Bitsch, F, Bourgier, E, Geiser, M, Hemmig, R, Kroemer, M, Lehmann, S, Ramage, P, Rieffel, S, Strauss, A, Green, J.R, Jahnke, W. | | Deposit date: | 2005-12-02 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the exceptional in vivo efficacy of bisphosphonate drugs.
Chemmedchem, 1, 2006
|
|
2IRW
 
 | | Human 11-beta-Hydroxysteroid Dehydrogenase (HSD1) with NADP and Adamantane Ether Inhibitor | | Descriptor: | (1S,3R,4S,5S,7S)-4-{[2-(4-METHOXYPHENOXY)-2-METHYLPROPANOYL]AMINO}ADAMANTANE-1-CARBOXAMIDE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Longenecker, K.L, Patel, J.R, Russell, J, Qin, W. | | Deposit date: | 2006-10-16 | | Release date: | 2007-01-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of adamantane ethers as inhibitors of 11beta-HSD-1: Synthesis and biological evaluation.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5U7M
 
 | | Crystal Structure of HIV-1 BG505 SOSIP.664 Prefusion Env Trimer Bound to Small Molecule HIV-1 Entry Inhibitor BMS-378806 in Complex with Human Antibodies PGT122 and 35O22 at 3.8 Angstrom | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pancera, M, Lai, Y.-T, Kwong, P.D. | | Deposit date: | 2016-12-12 | | Release date: | 2017-08-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.025 Å) | | Cite: | Crystal structures of trimeric HIV envelope with entry inhibitors BMS-378806 and BMS-626529.
Nat. Chem. Biol., 13, 2017
|
|
5UTY
 
 | | Crystal Structure of a Stabilized DS-SOSIP.mut4 BG505 gp140 HIV-1 Env Trimer, Containing Mutations I201C-P433C (DS), L154M, N300M, N302M, T320L in Complex with Human Antibodies PGT122 and 35O22 at 4.1 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Fab heavy chain, ... | | Authors: | Xu, K, Chuang, G.-Y, Pancera, M, Kwong, P.D. | | Deposit date: | 2017-02-15 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.412 Å) | | Cite: | Structure-Based Design of a Soluble Prefusion-Closed HIV-1 Env Trimer with Reduced CD4 Affinity and Improved Immunogenicity.
J. Virol., 91, 2017
|
|
3N8Y
 
 | | Structure of Aspirin Acetylated Cyclooxygenase-1 in Complex with Diclofenac | | Descriptor: | 2-HYDROXYBENZOIC ACID, 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sidhu, R.S. | | Deposit date: | 2010-05-28 | | Release date: | 2010-07-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Comparison of Cyclooxygenase-1 Crystal Structures: Cross-Talk between Monomers Comprising Cyclooxygenase-1 Homodimers
Biochemistry, 49, 2010
|
|
3N8W
 
 | |
