9C1N
 
 | | HerA-DUF4297 assembly 2 | | Descriptor: | ATP-binding protein, DUF4297 domain-containing protein | | Authors: | Rish, A.D, Fosuah, E, Fu, T.M. | | Deposit date: | 2024-05-29 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Architecture remodeling activates the HerA-DUF anti-phage defense system.
Mol.Cell, 85, 2025
|
|
9C1X
 
 | | Apo DUF4297 12-mer | | Descriptor: | DUF4297 domain-containing protein | | Authors: | Rish, A.D, Fosuah, E, Fu, T.M. | | Deposit date: | 2024-05-29 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Architecture remodeling activates the HerA-DUF anti-phage defense system.
Mol.Cell, 85, 2025
|
|
9C1O
 
 | |
9C5X
 
 | |
9C1M
 
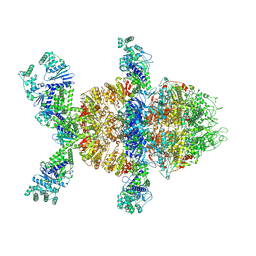 | | HerA-DUF assembly 1 | | Descriptor: | ATP-binding protein, DUF4297 domain-containing protein | | Authors: | Rish, A.D, Fosuah, E, Fu, T.M. | | Deposit date: | 2024-05-29 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Architecture remodeling activates the HerA-DUF anti-phage defense system.
Mol.Cell, 85, 2025
|
|
6Q3X
 
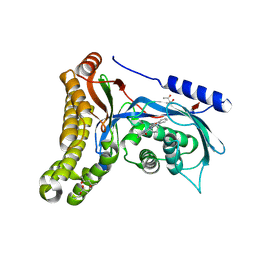 | | Structure of human galactokinase in complex with galactose and 2'-(benzo[d]oxazol-2-ylamino)-7',8'-dihydro-1'H-spiro[cyclohexane-1,4'-quinazolin]-5'(6'H)-one | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, ... | | Authors: | Bezerra, G.A, Mackinnon, S, Zhang, M, Foster, W, Bailey, H, Arrowsmith, C, Edwards, A, Bountra, C, Lai, K, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-04 | | Release date: | 2020-07-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
6GGS
 
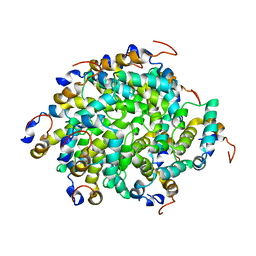 | | Structure of RIP2 CARD filament | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Pellegrini, E, Cusack, S, Desfosses, A, Schoehn, G, Malet, H, Gutsche, I, Sachse, C, Hons, M. | | Deposit date: | 2018-05-03 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | RIP2 filament formation is required for NOD2 dependent NF-kappa B signalling.
Nat Commun, 9, 2018
|
|
5IPE
 
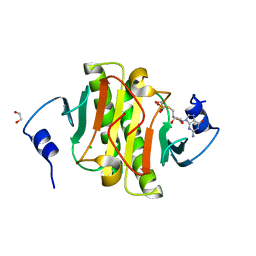 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) nucleoside thiophosphoramidate catalytic product complex | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-phosphono-5'-thioguanosine, CHLORIDE ION, ... | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Caught before Released: Structural Mapping of the Reaction Trajectory for the Sofosbuvir Activating Enzyme, Human Histidine Triad Nucleotide Binding Protein 1 (hHint1).
Biochemistry, 56, 2017
|
|
5IPB
 
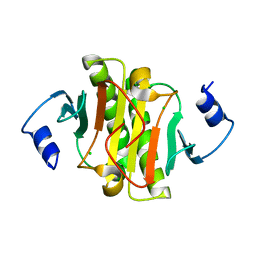 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant | | Descriptor: | CHLORIDE ION, Histidine triad nucleotide-binding protein 1 | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Caught before Released: Structural Mapping of the Reaction Trajectory for the Sofosbuvir Activating Enzyme, Human Histidine Triad Nucleotide Binding Protein 1 (hHint1).
Biochemistry, 56, 2017
|
|
6GK2
 
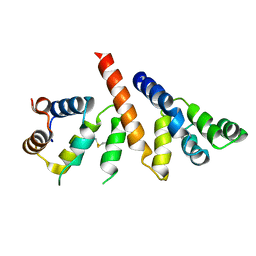 | | Helical reconstruction of BCL10 CARD and MALT1 DEATH DOMAIN complex | | Descriptor: | B-cell lymphoma/leukemia 10, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Schlauderer, F, Desfosses, A, Gutsche, I, Hopfner, K.P, Lammens, K. | | Deposit date: | 2018-05-18 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Molecular architecture and regulation of BCL10-MALT1 filaments.
Nat Commun, 9, 2018
|
|
5IPC
 
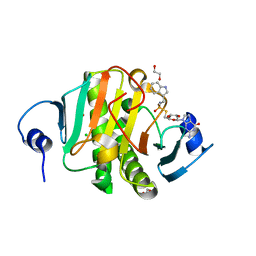 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant nucleoside thiophosphoramidate substrate complex | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-[(S)-hydroxy{[2-(1H-indol-3-yl)ethyl]amino}phosphoryl]-5'-thioguanosine, CHLORIDE ION, ... | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Caught before Released: Structural Mapping of the Reaction Trajectory for the Sofosbuvir Activating Enzyme, Human Histidine Triad Nucleotide Binding Protein 1 (hHint1).
Biochemistry, 56, 2017
|
|
5IPD
 
 | |
9E0O
 
 | | CryoEM structure of inducible Lysine decarboxylase from Hafnia alvei L-hydrazino-Lysine analog at 2.04 Angstrom resolution | | Descriptor: | (2R)-6-amino-2-[(2E)-2-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)hydrazin-1-yl]hexanoic acid, Lysine decarboxylase, inducible | | Authors: | Duhoo, Y, Desfosses, A, Gutsche, I, Doukov, T.I, Berkowitz, D.B. | | Deposit date: | 2024-10-18 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | alpha-Hydrazino Acids Inhibit Pyridoxal Phosphate-Dependent Decarboxylases via "Catalytically Correct" Ketoenamine Tautomers: A Special Motif for Chemical Biology and Drug Discovery?
Acs Catalysis, 15, 2025
|
|
9E0M
 
 | | CryoEM structure of holoenzyme of inducible Lysine decarboxylase from Hafnia alvei holoenzyme at 2.19 Angstrom resolution | | Descriptor: | Lysine decarboxylase, inducible | | Authors: | Duhoo, Y, Desfosses, A, Gutsche, I, Doukov, T.I, Berkowitz, D.B. | | Deposit date: | 2024-10-18 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | alpha-Hydrazino Acids Inhibit Pyridoxal Phosphate-Dependent Decarboxylases via "Catalytically Correct" Ketoenamine Tautomers: A Special Motif for Chemical Biology and Drug Discovery?
Acs Catalysis, 15, 2025
|
|
6OFU
 
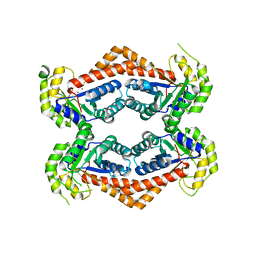 | | X-ray crystal structure of the YdjI aldolase from Escherichia coli K12 | | Descriptor: | CHLORIDE ION, YdjI aldolase, ZINC ION | | Authors: | Dopkins, B.J, Thoden, J.B, Huddleston, J.P, Narindoshvili, T, Fose, B, Rachel, F.M, Holden, H.M. | | Deposit date: | 2019-04-01 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Functional Characterization of YdjI, an Aldolase of Unknown Specificity inEscherichia coliK12.
Biochemistry, 58, 2019
|
|
2OXO
 
 | |
8BD7
 
 | | IFTB1 subcomplex of anterograde Intraflagellar transport trains (Chlamydomonas reinhardtii) | | Descriptor: | Clusterin-associated protein 1, IFT54, IFT70, ... | | Authors: | Lacey, S.E, Foster, H.E, Pigino, G. | | Deposit date: | 2022-10-18 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (9.9 Å) | | Cite: | The molecular structure of IFT-A and IFT-B in anterograde intraflagellar transport trains.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BDA
 
 | | IFTA complex in anterograde intraflagellar transport trains (Chlamydomonas reinhardtii) | | Descriptor: | Intraflagellar transport particle protein 140, Intraflagellar transport protein 121, Intraflagellar transport protein 122 homolog, ... | | Authors: | Lacey, S.E, Foster, H.E, Pigino, G. | | Deposit date: | 2022-10-18 | | Release date: | 2023-01-11 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (20.700001 Å) | | Cite: | The molecular structure of IFT-A and IFT-B in anterograde intraflagellar transport trains.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AXB
 
 | | Cryo-EM structure of Cas12k-sgRNA binary complex (type V-K CRISPR-associated transposon) | | Descriptor: | Cas12k, sgRNA | | Authors: | Tenjo-Castano, F, Sofos, N, Stella, S, Molina, R, Pape, T, Lopez-Mendez, B, Stutzke, L.S, Temperini, P, Montoya, G. | | Deposit date: | 2022-08-31 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Conformational landscape of the type V-K CRISPR-associated transposon integration assembly.
Mol.Cell, 84, 2024
|
|
8AXA
 
 | | Cryo-EM structure of shCas12k-sgRNA-dsDNA ternary complex (type V-K CRISPR-associated transposon) | | Descriptor: | Cas12k, DNA non-target strand, DNA target strand, ... | | Authors: | Tenjo-Castano, F, Sofos, N, Stella, S, Fuglsang, A, Pape, T, Mesa, P, Stutzke, L.S, Temperini, P, Montoya, G. | | Deposit date: | 2022-08-31 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Conformational landscape of the type V-K CRISPR-associated transposon integration assembly.
Mol.Cell, 84, 2024
|
|
3MYY
 
 | | Structure of E. Coli CheY mutant A113P bound to Beryllium fluoride | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein cheY, GLYCEROL, ... | | Authors: | Immormino, R.M, McDonald, L.R, Bourret, R.B. | | Deposit date: | 2010-05-11 | | Release date: | 2011-05-11 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of Position K+4 in the Phosphorylation and Dephosphorylation Reaction Kinetics of the CheY Response Regulator.
Biochemistry, 60, 2021
|
|
3AU0
 
 | | Structural and biochemical characterization of ClfB:ligand interactions | | Descriptor: | Clumping factor B, MAGNESIUM ION | | Authors: | Ganesh, V.K, Barbu, E.M, Deivanayagam, C.C.S, Le, B, Anderson, A.S, Matsuka, Y, Lin, S.L, Foster, T.F, Narayana, S.V.L, Hook, M. | | Deposit date: | 2011-01-28 | | Release date: | 2011-05-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and biochemical characterization of ClfB:ligand interactions
To be published
|
|
2ZKJ
 
 | | Crystal structure of human PDK4-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Li, J, Chuang, D.T. | | Deposit date: | 2008-03-25 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pyruvate Dehydrogenase Kinase-4 Structures Reveal a Metastable Open Conformation Fostering Robust Core-free Basal Activity
J.Biol.Chem., 283, 2008
|
|
1SUL
 
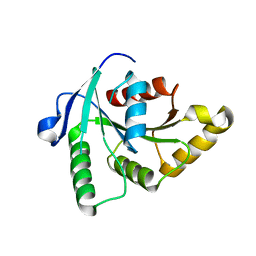 | | Crystal Structure of the apo-YsxC | | Descriptor: | GTP-binding protein YsxC | | Authors: | Ruzheinikov, S.N, Das, K.S, Sedelnikova, S.E, Baker, P.J, Artymiuk, P.J, Garcia-Lara, J, Foster, S.J, Rice, D.W. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of the Open and Closed Conformations of the GTP-binding Protein YsxC from Bacillus subtilis.
J.Mol.Biol., 339, 2004
|
|
1SVI
 
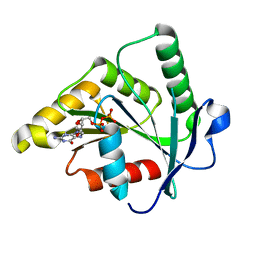 | | Crystal Structure of the GTP-binding protein YsxC complexed with GDP | | Descriptor: | GTP-binding protein YSXC, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Ruzheinikov, S.N, Das, S.K, Sedelnikova, S.E, Baker, P.J, Artymiuk, P.J, Garcia-Lara, J, Foster, S.J, Rice, D.W. | | Deposit date: | 2004-03-29 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Analysis of the Open and Closed Conformations of the GTP-binding Protein YsxC from Bacillus subtilis.
J.Mol.Biol., 339, 2004
|
|
