2BAI
 
 | | The Zinc finger domain of Mengovirus Leader polypeptide | | Descriptor: | Genome polyprotein, ZINC ION | | Authors: | Cornilescu, C.C, Porter, F.W, Qin, Z, Lee, M.S, Palmenberg, A.C, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-14 | | Release date: | 2006-01-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the mengovirus Leader protein zinc-finger domain.
Febs Lett., 582, 2008
|
|
3V7S
 
 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with inhibitor 0364 | | Descriptor: | 5-methyl-3-[4-(4-{5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentyl}-1H-1,2,3-triazol-1-yl)butyl]-1,3-benzoxazol-2(3H)-one, Biotin ligase | | Authors: | Yap, M.Y, Pendini, N.R. | | Deposit date: | 2011-12-21 | | Release date: | 2012-04-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Selective inhibition of biotin protein ligase from Staphylococcus aureus.
J.Biol.Chem., 287, 2012
|
|
2AYX
 
 | | Solution structure of the E.coli RcsC C-terminus (residues 700-949) containing linker region and phosphoreceiver domain | | Descriptor: | Sensor kinase protein rcsC | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Koglin, A, Lohr, F, Dotsch, V. | | Deposit date: | 2005-09-09 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A New Structural Domain in the Escherichia coli RcsC Hybrid Sensor Kinase Connects Histidine Kinase and Phosphoreceiver Domains
J.Mol.Biol., 364, 2006
|
|
3VE4
 
 | |
1ZHO
 
 | | The structure of a ribosomal protein L1 in complex with mRNA | | Descriptor: | 50S ribosomal protein L1, POTASSIUM ION, mRNA | | Authors: | Nevskaya, N, Tishchenko, S, Volchkov, S, Kljashtorny, V, Nikonova, E, Nikonov, O, Nikulin, A, Kohrer, C, Piendl, W, Zimmermann, R, Stockley, P, Garber, M, Nikonov, S. | | Deposit date: | 2005-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New insights into the interaction of ribosomal protein L1 with RNA.
J.Mol.Biol., 355, 2006
|
|
1ZB6
 
 | | Co-Crystal Structure of ORF2 an Aromatic Prenyl Transferase from Streptomyces sp. strain cl190 complexed with GSPP and 1,6-dihydroxynaphtalene | | Descriptor: | 1,6-DIHYDROXY NAPHTHALENE, Aromatic prenyltransferase, GERANYL S-THIOLODIPHOSPHATE, ... | | Authors: | Kuzuyama, T, Noel, J.P, Richard, S.B. | | Deposit date: | 2005-04-07 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products.
Nature, 435, 2005
|
|
2B43
 
 | |
2AGH
 
 | | Structural basis for cooperative transcription factor binding to the CBP coactivator | | Descriptor: | Crebbp protein, Myb proto-oncogene protein, Zinc finger protein HRX | | Authors: | De Guzman, R.N, Goto, N.K, Dyson, H.J, Wright, P.E. | | Deposit date: | 2005-07-26 | | Release date: | 2005-11-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Cooperative Transcription Factor Binding to the CBP Coactivator
J.Mol.Biol., 355, 2006
|
|
1XGI
 
 | | AmpC beta-lactamase in complex with 3-(3-nitro-phenylsulfamoyl)-thiophene-2-carboxylic acid | | Descriptor: | 3-{[(3-NITROANILINE]SULFONYL}THIOPHENE-2-CARBOXYLIC ACID, Beta-lactamase | | Authors: | Tondi, D, Morandi, F, Bonnet, R, Costi, M.P, Shoichet, B.K. | | Deposit date: | 2004-09-17 | | Release date: | 2005-05-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-based optimization of a non-beta-lactam lead results in inhibitors that do not up-regulate beta-lactamase expression in cell culture.
J.Am.Chem.Soc., 127, 2005
|
|
2O25
 
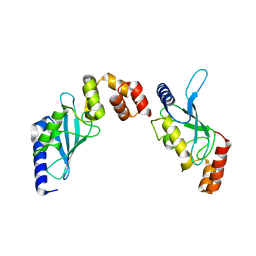 | | Ubiquitin-Conjugating Enzyme E2-25 kDa Complexed With SUMO-1-Conjugating Enzyme UBC9 | | Descriptor: | SUMO-1-conjugating enzyme UBC9, Ubiquitin-conjugating enzyme E2-25 kDa | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-29 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Novel and Unexpected Complex Between the SUMO-1-Conjugating Enzyme UBC9 and the Ubiquitin-Conjugating Enzyme E2-25 kDa
To be Published
|
|
1XFP
 
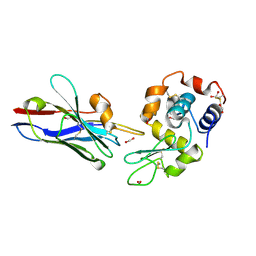 | | Crystal structure of the CDR2 germline reversion mutant of cAb-Lys3 in complex with hen egg white lysozyme | | Descriptor: | FORMIC ACID, Lysozyme C, heavy chain antibody | | Authors: | De Genst, E, Handelberg, F, Van Meirhaeghe, A, Vynck, S, Loris, R, Wyns, L, Muyldermans, S. | | Deposit date: | 2004-09-15 | | Release date: | 2004-09-28 | | Last modified: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Chemical Basis for the Affinity Maturation of a Camel Single Domain Antibody
J.Biol.Chem., 279, 2004
|
|
3D97
 
 | |
1G1X
 
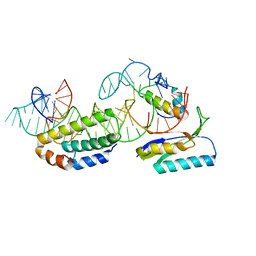 | | STRUCTURE OF RIBOSOMAL PROTEINS S15, S6, S18, AND 16S RIBOSOMAL RNA | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S15, 30S RIBOSOMAL PROTEIN S18, ... | | Authors: | Agalarov, S.C, Prasad, G.S, Funke, P.M, Stout, C.D, Williamson, J.R. | | Deposit date: | 2000-10-13 | | Release date: | 2000-10-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the S15,S6,S18-rRNA complex: assembly of the 30S ribosome central domain.
Science, 288, 2000
|
|
1G2M
 
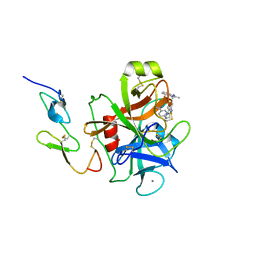 | | FACTOR XA INHIBITOR COMPLEX | | Descriptor: | 4-{[1-METHYL-5-(2-METHYL-BENZOIMIDAZOL-1-YLMETHYL)-1H-BENZOIMIDAZOL-2-YLMETHYL]-AMINO}-BENZAMIDINE, CALCIUM ION, COAGULATION FACTOR X | | Authors: | Nar, H. | | Deposit date: | 2000-10-20 | | Release date: | 2001-10-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural basis for inhibition promiscuity of dual specific thrombin and factor Xa blood coagulation inhibitors.
Structure, 9, 2001
|
|
1XTK
 
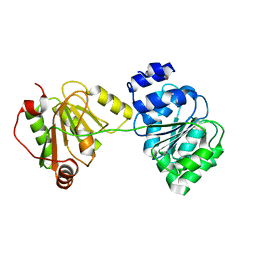 | | structure of DECD to DEAD mutation of human UAP56 | | Descriptor: | BETA-MERCAPTOETHANOL, Probable ATP-dependent RNA helicase p47 | | Authors: | Shi, H, Cordin, O, Minder, C.M, Linder, P, Xu, R.M. | | Deposit date: | 2004-10-22 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human ATP-dependent splicing and export factor UAP56
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1Y39
 
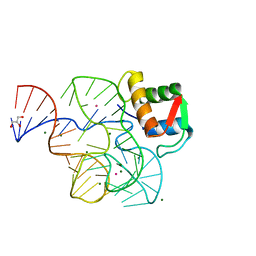 | | Co-evolution of protein and RNA structures within a highly conserved ribosomal domain | | Descriptor: | 50S ribosomal protein L11, 58 Nucleotide Ribosomal 23S RNA Domain, COBALT (III) ION, ... | | Authors: | Dunstan, M.S, GuhaThakurta, D, Draper, D.E, Conn, G.L. | | Deposit date: | 2004-11-24 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Coevolution of Protein and RNA Structures within a Highly Conserved Ribosomal Domain
Chem.Biol., 12, 2005
|
|
5CR4
 
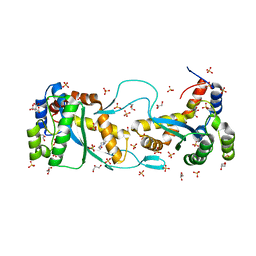 | |
3DM1
 
 | | Crystal structure of the complex of human chromobox homolog 3 (CBX3) with peptide | | Descriptor: | Chromobox protein homolog 3, Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 | | Authors: | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-19 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of the chromodomain of Cbx3 bound to methylated peptides from histone h1 and G9a.
Plos One, 7, 2012
|
|
1Y6S
 
 | | HIV-1 DIS(Mal) duplex Ba-soaked | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', BARIUM ION | | Authors: | Ennifar, E, Walter, P, Dumas, P. | | Deposit date: | 2004-12-07 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A crystallographic study of the binding of 13 metal ions to two related RNA duplexes
Nucleic Acids Res., 31, 2003
|
|
2EJR
 
 | | LSD1-tranylcypromine complex | | Descriptor: | Lysine-specific histone demethylase 1, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL (2R,3S,4S)-5-[7,8-DIMETHYL-2,4-DIOXO-5-(3-PHENYLPROPANOYL)-1,3,4,5-TETRAHYDROBENZO[G]PTERIDIN-10(2H)-YL]-2,3,4-TRIHYDROXYPENTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Sengoku, T, Mimasu, S, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-20 | | Release date: | 2008-01-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of histone demethylase LSD1 and tranylcypromine at 2.25A
Biochem.Biophys.Res.Commun., 366, 2008
|
|
2EL6
 
 | |
2EL5
 
 | |
5DGF
 
 | | Complex of yeast 80S ribosome with hypusine-containing/non-modified eIF5A and/or a peptidyl-tRNA analog | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Melnikov, S, Mailliot, J, Shin, B.-S, Rigger, L, Yusupova, G, Micura, R, Dever, T.E, Yusupov, M. | | Deposit date: | 2015-08-27 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Coping with proline stalling: structural basis of hypusine-induced protein synthesis by the eukaryotic ribosome
To Be Published
|
|
3SFG
 
 | |
1Y95
 
 | | HIV-1 Dis(Mal) Duplex Pb-Soaked | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', LEAD (II) ION, MAGNESIUM ION | | Authors: | Ennifar, E, Walter, P, Dumas, P. | | Deposit date: | 2004-12-14 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A crystallographic study of the binding of 13 metal ions to two related RNA duplexes
Nucleic Acids Res., 31, 2003
|
|
