1UU3
 
 | | Structure of human PDK1 kinase domain in complex with LY333531 | | Descriptor: | (9R)-9-[(DIMETHYLAMINO)METHYL]-6,7,10,11-TETRAHYDRO-9H,18H-5,21:12,17-DIMETHENODIBENZO[E,K]PYRROLO[3,4-H][1,4,13]OXADIA ZACYCLOHEXADECINE-18,20-DIONE, 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE-1, GLYCEROL, ... | | Authors: | Komander, D, Kular, G.S, Schuttelkopf, A.W, Deak, M, Prakash, K.R, Bain, J, Elliot, M, Garrido-Franco, M, Kozikowski, A.P, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2003-12-15 | | Release date: | 2004-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interactions of Ly333531 and Other Bisindolyl Maleimide Inhibitors with Pdk1
Structure, 12, 2004
|
|
1HL7
 
 | | Gamma lactamase from an Aureobacterium species in complex with 3a,4,7,7a-tetrahydro-benzo [1,3] dioxol-2-one | | Descriptor: | 3A,4,7,7A-TETRAHYDRO-BENZO [1,3] DIOXOL-2-ONE, GAMMA LACTAMASE | | Authors: | Line, K, Isupov, M.N, Littlechild, J.A. | | Deposit date: | 2003-03-14 | | Release date: | 2004-03-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Crystal Structure of a (-)Gamma-Lactamase from an Aureobacterium Species Reveals a Tetrahedral Intermediate in the Active Site
J.Mol.Biol., 338, 2004
|
|
1DQ8
 
 | | COMPLEX OF THE CATALYTIC PORTION OF HUMAN HMG-COA REDUCTASE WITH HMG AND COA | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-HYDROXY-3-METHYL-GLUTARIC ACID, COENZYME A, ... | | Authors: | Istvan, E.S, Palnitkar, M, Buchanan, S.K, Deisenhofer, J. | | Deposit date: | 1999-12-30 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis.
EMBO J., 19, 2000
|
|
1UZU
 
 | | Glycogen Phosphorylase b in complex with indirubin-5'-sulphonate | | Descriptor: | 2',3-DIOXO-1,1',2',3-TETRAHYDRO-2,3'-BIINDOLE-5'-SULFONIC ACID, GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, ... | | Authors: | Kosmopoulou, M.N, Leonidas, D.D, Chrysina, E.D, Bischler, N, Eisenbrand, G, Sakarellos, C.E, Pauptit, R, Oikonomakos, N.G. | | Deposit date: | 2004-03-16 | | Release date: | 2004-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding of the potential antitumour agent indirubin-5-sulphonate at the inhibitor site of rabbit muscle glycogen phosphorylase b. Comparison with ligand binding to pCDK2-cyclin A complex.
Eur. J. Biochem., 271, 2004
|
|
1TCW
 
 | | SIV PROTEASE COMPLEXED WITH INHIBITOR SB203386 | | Descriptor: | (2R,4S,5S,1'S)-2-PHENYLMETHYL-4-HYDROXY-5-(TERT-BUTOXYCARBONYL)AMINO-6-PHENYL HEXANOYL-N-(1'-IMIDAZO-2-YL)-2'-METHYLPROPANAMIDE, SIV PROTEASE | | Authors: | Hoog, S.S, Abdel-Meguid, S.S. | | Deposit date: | 1996-06-05 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human immunodeficiency virus protease ligand specificity conferred by residues outside of the active site cavity.
Biochemistry, 35, 1996
|
|
3AIQ
 
 | | Crystal structure of beta-glucosidase in wheat complexed with an aglycone DIMBOA | | Descriptor: | 2,4-DIHYDROXY-7-(METHYLOXY)-2H-1,4-BENZOXAZIN-3(4H)-ONE, Beta-glucosidase | | Authors: | Sue, M, Nakamura, C, Miyamoto, T, Yajima, S. | | Deposit date: | 2010-05-18 | | Release date: | 2011-02-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active-site architecture of benzoxazinone-glucoside beta-D-glucosidases in Triticeae
Plant Sci., 180, 2011
|
|
2HCR
 
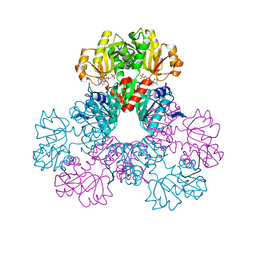 | | crystal structure of human phosphoribosyl pyrophosphate synthetase 1 in complex with AMP(ATP), cadmium and sulfate ion | | Descriptor: | ADENOSINE MONOPHOSPHATE, CADMIUM ION, Ribose-phosphate pyrophosphokinase I, ... | | Authors: | Li, S, Peng, B, Ding, J. | | Deposit date: | 2006-06-18 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human phosphoribosylpyrophosphate synthetase 1 reveals a novel allosteric site
Biochem.J., 401, 2007
|
|
3AHA
 
 | | Crystal structure of the complex between gp41 fragments N36 and C34 mutant N126K/E137Q | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Transmembrane protein gp41 | | Authors: | Izumi, K, Nakamura, S, Nakano, H, Shimura, K, Sakagami, Y, Oishi, S, Uchiyama, S, Ohkubo, T, Kobayashi, Y, Fujii, N, Matsuoka, M, Kodama, E.N. | | Deposit date: | 2010-04-22 | | Release date: | 2010-05-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of HIV-1 resistance to a fusion inhibitor, N36, derived from the gp41 amino terminal heptad repeat.
Antiviral Res., 2010
|
|
1SUP
 
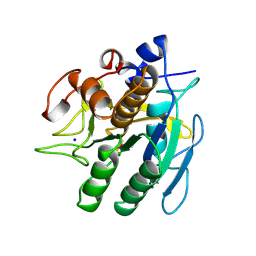 | | SUBTILISIN BPN' AT 1.6 ANGSTROMS RESOLUTION: ANALYSIS OF DISCRETE DISORDER AND COMPARISON OF CRYSTAL FORMS | | Descriptor: | CALCIUM ION, SODIUM ION, SUBTILISIN BPN', ... | | Authors: | Gallagher, D.T, Oliver, J.D, Betzel, C, Gilliland, G.L. | | Deposit date: | 1995-08-14 | | Release date: | 1995-11-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Subtilisin BPN' at 1.6 A resolution: analysis for discrete disorder and comparison of crystal forms.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
4PMI
 
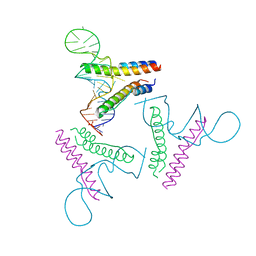 | | Crystal structure of Rev and Rev-response-element RNA complex | | Descriptor: | PHOSPHATE ION, Protein Rev, Rev-Response-Element RNA | | Authors: | Jayaraman, B, Crosby, D.C, Homer, C, Ribeiro, I, Mavor, D, Frankel, A.D. | | Deposit date: | 2014-05-21 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | RNA-directed remodeling of the HIV-1 protein Rev orchestrates assembly of the Rev-Rev response element complex.
Elife, 4, 2014
|
|
3OOF
 
 | |
1MXG
 
 | | Crystal Structure of a (Ca,Zn)-dependent alpha-amylase from the hyperthermophilic archaeon Pyrococcus woesei in complex with acarbose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Linden, A, Mayans, O, Meyer-Klaucke, W, Antranikian, G, Wilmanns, M. | | Deposit date: | 2002-10-02 | | Release date: | 2003-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Differential Regulation of a Hyperthermophilic alpha-Amylase with a Novel (Ca,Zn) Two-metal Center by Zinc
J.Biol.Chem., 278, 2003
|
|
3INQ
 
 | | Crystal structure of BCL-XL in complex with W1191542 | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-(biphenyl-3-ylmethyl)piperazin-1-yl]-N-{[4-({(1R)-3-(dimethylamino)-1-[(phenylsulfanyl)methyl]propyl}amino)-3-nitrophenyl]sulfonyl}benzamide, Bcl-2-like protein 1, ... | | Authors: | Fairlie, W.D, Smith, B.J, Colman, P.M, Czabotar, P.E, Lee, E.F. | | Deposit date: | 2009-08-12 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands
J. Biol. Chem., 284, 2009
|
|
5VTX
 
 | |
1TUK
 
 | | Crystal structure of liganded type 2 non specific lipid transfer protein from wheat | | Descriptor: | 1-MYRISTOYL-2-HYDROXY-SN-GLYCERO-3-[PHOSPHO-RAC-(1-GLYCEROL)], IODIDE ION, Nonspecific lipid-transfer protein 2G | | Authors: | Hoh, F, Pons, J.L, Gautier, M.F, De Lamotte, F, Dumas, C. | | Deposit date: | 2004-06-25 | | Release date: | 2005-04-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structure of a liganded type 2 non-specific lipid-transfer protein from wheat and the molecular basis of lipid binding.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5REM
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0103016 | | Descriptor: | 1 1-(4-(2-nitrophenyl)piperazin-1-yl)ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
1MVY
 
 | | Amylosucrase mutant E328Q co-crystallized with maltoheptaose. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Skov, L.K, Mirza, O, Sprogoe, D, Dar, I, Remaud-Simeon, M, Albenne, C, Monsan, P, Gajhede, M. | | Deposit date: | 2002-09-27 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide and Sucrose Complexes of Amylosucrase. STRUCTURAL IMPLICATIONS FOR THE POLYMERASE ACTIVITY
J.BIOL.CHEM., 277, 2002
|
|
3IHU
 
 | |
1TSU
 
 | | CRYSTAL STRUCTURE OF DECAMER NCP1 SUBSTRATE PEPTIDE IN COMPLEX WITH WILD-TYPE D25N HIV-1 PROTEASE VARIANT | | Descriptor: | NC-P1 SUBSTRATE PEPTIDE, Pol polyprotein | | Authors: | Prabu-Jeyabalan, M, Nalivaika, E.A, King, N.M, Schiffer, C.A. | | Deposit date: | 2004-06-21 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for coevolution of a human immunodeficiency virus type 1 nucleocapsid-p1 cleavage site with a V82A drug-resistant mutation in viral protease
J.Virol., 78, 2004
|
|
7E3H
 
 | | Crystal structure of human acetylcholinesterase in complex with donepezil | | Descriptor: | 1-BENZYL-4-[(5,6-DIMETHOXY-1-INDANON-2-YL)METHYL]PIPERIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Dileep, K.V, Ihara, K, Mishima-Tsumagari, C, Kukimoto-Niino, M, Yonemochi, M, Hanada, K, Shirouzu, M, Zhang, K.Y.J. | | Deposit date: | 2021-02-08 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of human acetylcholinesterase in complex with tacrine: Implications for drug discovery
Int.J.Biol.Macromol., 210, 2022
|
|
2WT2
 
 | | Galectin domain of porcine adenovirus type 4 NADC-1 isolate fibre complexed with tri(N-acetyl-lactosamine) | | Descriptor: | NITRATE ION, PUTATIVE FIBER PROTEIN, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Guardado-Calvo, P, Munoz, E.M, Llamas-Saiz, A.L, Fox, G.C, Glasgow, J.N, van Raaij, M.J. | | Deposit date: | 2009-09-10 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic structure of porcine adenovirus type 4 fiber head and galectin domains.
J. Virol., 84, 2010
|
|
3OIL
 
 | | Human Carbonic anhydrase II mutant A65S, N67Q (CA IX mimic) bound by 2-Ethylestradiol 3-O-sulfamate | | Descriptor: | (14beta,17alpha)-2-ethyl-17-hydroxyestra-1(10),2,4-trien-3-yl sulfamate, Carbonic anhydrase 2, ZINC ION | | Authors: | Sippel, K.H, Stander, B.A, Robbins, A.H, Tu, C.K, Agbandje-McKenna, M, Silverman, D.N, Joubert, A.M, McKenna, R. | | Deposit date: | 2010-08-19 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization of Carbonic Anhydrase Isozyme Specific Inhibition by Sulfamated 2-Ethylestra Compounds
LETT.DRUG DES.DISCOVERY, 8, 2011
|
|
5RY0
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z275165822 | | Descriptor: | 1-[4-(4-chlorophenyl)piperazin-1-yl]ethan-1-one, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
4OIA
 
 | | Crystal Structure of ICAM-5 D1-D4 ectodomain fragment, Space Group P4322 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Recacha, R, Jimenez, D, Tian, L, Barredo, R, Ghamberg, C, Casasnovas, J.M. | | Deposit date: | 2014-01-19 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structures of an ICAM-5 ectodomain fragment show electrostatic-based homophilic adhesions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HKL
 
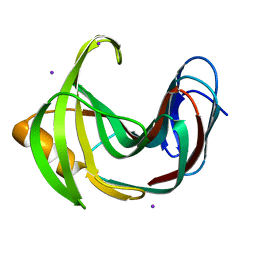 | | Crystal Structures of Mutant Endo-beta-1,4-xylanase II Complexed with substrate (1.15 A) and Products (1.6 A) | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Langan, P, Wan, Q, Coates, L, Kovalevsky, A. | | Deposit date: | 2012-10-15 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
