4I3X
 
 | |
3DMT
 
 | | Structure of Glycosomal Glyceraldehyde-3-Phosphate Dehydrogenase from Trypanosoma cruzi in complex with the irreversible iodoacetate inhibitor | | Descriptor: | GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, glycosomal, ... | | Authors: | Guido, R.V.C, Balliano, T.L, Andricopulo, A.D, Oliva, G. | | Deposit date: | 2008-07-01 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and Crystallographic Studies on Glyceraldehyde-3-Phosphate Dehydrogenase from Trypanosoma cruzi in Complex with Iodoacetate.
Letters in drug design & discovery, 6, 2009
|
|
4QO7
 
 | |
6JL0
 
 | | Crystal structure of VvPlpA from Vibrio vulnificus | | Descriptor: | BROMIDE ION, Thermolabile hemolysin | | Authors: | Ma, Q, Wan, Y, Liu, C. | | Deposit date: | 2019-03-03 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.073 Å) | | Cite: | Structural analysis of aVibriophospholipase reveals an unusual Ser-His-chloride catalytic triad.
J.Biol.Chem., 294, 2019
|
|
1YQO
 
 | | T268A mutant heme domain of flavocytochrome P450 BM3 | | Descriptor: | Bifunctional P-450:NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Clark, J.P, Miles, C.S, Mowat, C.G, Walkinshaw, M.D, Reid, G.A, Daff, S.N, Chapman, S.K. | | Deposit date: | 2005-02-02 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of Thr268 and Phe393 in cytochrome P450 BM3.
J.Inorg.Biochem., 100, 2006
|
|
3HS6
 
 | | X-ray crystal structure of eicosapentaenoic acid bound to the cyclooxygenase channel of cyclooxygenase-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vecchio, A.J, Simmons, D.M, Malkowski, M.G. | | Deposit date: | 2009-06-10 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of fatty acid substrate binding to cyclooxygenase-2.
J.Biol.Chem., 285, 2010
|
|
2R6S
 
 | | Crystal structure of Gab protein | | Descriptor: | BICINE, FE (II) ION, GLYCEROL, ... | | Authors: | Lohkamp, B, Dobritzsch, D. | | Deposit date: | 2007-09-06 | | Release date: | 2008-06-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A mixture of fortunes: the curious determination of the structure of Escherichia coli BL21 Gab protein.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6AUK
 
 | |
4B4E
 
 | | 1.00 A Structure of Lysozyme Crystallized with (R)-2-methyl-2,4- pentanediol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CHLORIDE ION, LYSOZYME C | | Authors: | Jakoncic, J, Berger, J, Stauber, M, Axelbaum, A, Asherie, N. | | Deposit date: | 2012-07-30 | | Release date: | 2012-08-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystallization of Lysozyme with (R)-, (S)- and (Rs)-2-Methyl-2,4-Pentanediol
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6JLM
 
 | | XFEL structure of cyanobacterial photosystem II (dark state, dataset2) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
2RBR
 
 | |
6AX7
 
 | | The crystal structure of a lysyl hydroxylase from Acanthamoeba polyphaga mimivirus | | Descriptor: | FE (II) ION, Procollagen lysyl hydroxylase and glycosyltransferase | | Authors: | Guo, H, Tsai, C, Miller, M.D, Alvarado, S, Tainer, J.A, Phillips Jr, G.N, Kurie, J.M. | | Deposit date: | 2017-09-06 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Pro-metastatic collagen lysyl hydroxylase dimer assemblies stabilized by Fe2+-binding.
Nat Commun, 9, 2018
|
|
4I7L
 
 | | T4 Lysozyme L99A/M102H with phenol bound | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-11-30 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The impact of introducing a histidine into an apolar cavity site on docking and ligand recognition.
J.Med.Chem., 56, 2013
|
|
4QKX
 
 | | Structure of beta2 adrenoceptor bound to a covalent agonist and an engineered nanobody | | Descriptor: | 4-[(1R)-1-hydroxy-2-({2-[3-methoxy-4-(2-sulfanylethoxy)phenyl]ethyl}amino)ethyl]benzene-1,2-diol, Beta-2 adrenergic receptor, R9 protein, ... | | Authors: | Weichert, D, Kruse, A.C, Manglik, A, Hiller, C, Zhang, C, Huebner, H, Kobilka, B.K, Gmeiner, P. | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-23 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Covalent agonists for studying G protein-coupled receptor activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1YL7
 
 | | the crystal structure of Mycobacterium tuberculosis dihydrodipicolinate reductase (Rv2773c) in complex with NADH (crystal form C) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Dihydrodipicolinate reductase, MAGNESIUM ION | | Authors: | Janowski, R, Kefala, G, Weiss, M.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-01-19 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | The structure of dihydrodipicolinate reductase (DapB) from Mycobacterium tuberculosis in three crystal forms.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3DN6
 
 | | 1,3,5-trifluoro-2,4,6-trichlorobenzene binding in the hydrophobic cavity of T4 lysozyme L99A mutant | | Descriptor: | 1,3,5-trichloro-2,4,6-trifluorobenzene, 2-HYDROXYETHYL DISULFIDE, Lysozyme, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2008-07-01 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Halogenated benzenes bound within a non-polar cavity in T4 lysozyme provide examples of I...S and I...Se halogen-bonding.
J.Mol.Biol., 385, 2009
|
|
4QGZ
 
 | |
3DPC
 
 | |
4I5G
 
 | | Crystal structure of Ralstonia sp. alcohol dehydrogenase mutant N15G, G37D, R38V, R39S, A86N, S88A | | Descriptor: | Alclohol dehydrogenase/short-chain dehydrogenase | | Authors: | Jarasch, A, Lerchner, A, Meining, W, Schiefner, A, Skerra, A. | | Deposit date: | 2012-11-28 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic analysis and structure-guided engineering of NADPH-dependent Ralstonia sp. Alcohol dehydrogenase toward NADH cosubstrate specificity.
Biotechnol.Bioeng., 110, 2013
|
|
4QLJ
 
 | | Crystal structure of rice BGlu1 E386G/Y341A/Q187A mutant complexed with cellotetraose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, SULFATE ION, ... | | Authors: | Pengthaisong, S, Ketudat Cairns, J.R. | | Deposit date: | 2014-06-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Effects of active site cleft residues on oligosaccharide binding, hydrolysis, and glycosynthase activities of rice BGlu1 and its mutants
Protein Sci., 23, 2014
|
|
4QHB
 
 | |
6AY0
 
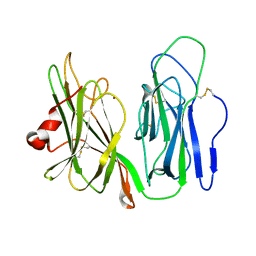 | | Crystal structure of H108A peptidylglycine alpha-hydroxylating monooxygenase (PHM) soaked with peptide | | Descriptor: | COPPER (II) ION, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-09-07 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
1YRZ
 
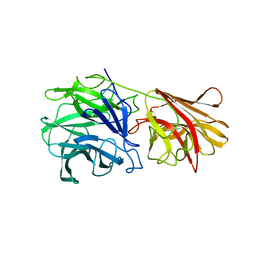 | | Crystal structure of xylan beta-1,4-xylosidase from Bacillus Halodurans C-125 | | Descriptor: | xylan beta-1,4-xylosidase | | Authors: | Fedorov, A.A, Fedorov, E.V, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-02-05 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of xylan beta-1,4-xylosidase from Bacillus Halodurans C-125
To be Published
|
|
4I6Y
 
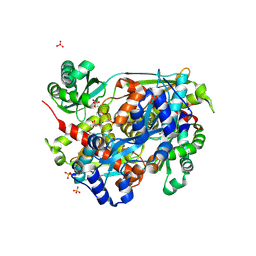 | | 3-hydroxy-3-methyl (HMG) Coenzyme A Reductase bound to R-Mevalonate | | Descriptor: | (R)-MEVALONATE, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, GLYCEROL, ... | | Authors: | Steussy, C.N, Stauffacher, C.V, Schmidt, T, Burgner II, J.W, Rodwell, V.W, Wrensford, L.V, Critchelow, C.J, Min, J. | | Deposit date: | 2012-11-30 | | Release date: | 2013-07-17 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Novel Role for Coenzyme A during Hydride Transfer in 3-Hydroxy-3-methylglutaryl-coenzyme A Reductase.
Biochemistry, 52, 2013
|
|
6JLK
 
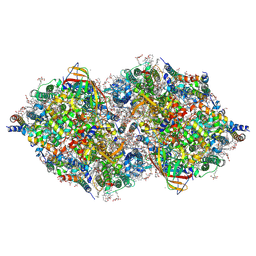 | | XFEL structure of cyanobacterial photosystem II (1F state, dataset1) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
