6PBK
 
 | |
6QNK
 
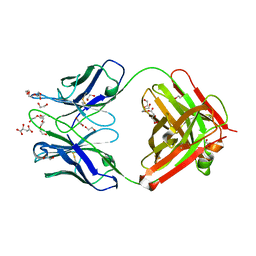 | | Antibody FAB fragment targeting Gi protein heterotrimer | | Descriptor: | 1,2-ETHANEDIOL, D-MALATE, FAB heavy chain, ... | | Authors: | Tsai, C.-J, Muehle, J, Pamula, F, Dawson, R.J.P, Maeda, S, Deupi, X, Schertler, G.F.X. | | Deposit date: | 2019-02-11 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cryo-EM structure of the rhodopsin-G alpha i-beta gamma complex reveals binding of the rhodopsin C-terminal tail to the G beta subunit.
Elife, 8, 2019
|
|
6QNO
 
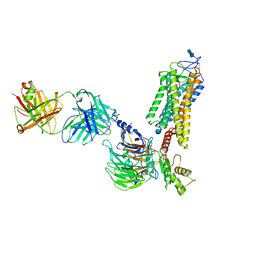 | | Rhodopsin-Gi protein complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab antibody fragment heavy chain, Fab antibody fragment light chain, ... | | Authors: | Tsai, C.-J, Marino, J, Adaixo, R.J, Pamula, F, Muehle, J, Maeda, S, Flock, T, Taylor, N.M.I, Mohammed, I, Matile, H, Dawson, R.J.P, Deupi, X, Stahlberg, H, Schertler, G.F.X. | | Deposit date: | 2019-02-11 | | Release date: | 2019-07-10 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.38 Å) | | Cite: | Cryo-EM structure of the rhodopsin-G alpha i-beta gamma complex reveals binding of the rhodopsin C-terminal tail to the G beta subunit.
Elife, 8, 2019
|
|
6FUF
 
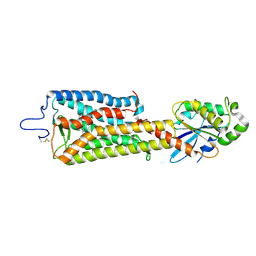 | | Crystal structure of the rhodopsin-mini-Go complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(o) subunit alpha, RETINAL, ... | | Authors: | Tsai, C.-J, Weinert, T, Muehle, J, Pamula, F, Nehme, R, Flock, T, Nogly, P, Edwards, P.C, Carpenter, B, Gruhl, T, Ma, P, Deupi, X, Standfuss, J, Tate, C.G, Schertler, G.F.X. | | Deposit date: | 2018-02-27 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.117 Å) | | Cite: | Crystal structure of rhodopsin in complex with a mini-Gosheds light on the principles of G protein selectivity.
Sci Adv, 4, 2018
|
|
4BGN
 
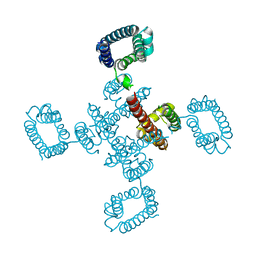 | | cryo-EM structure of the NavCt voltage-gated sodium channel | | Descriptor: | VOLTAGE-GATED SODIUM CHANNEL | | Authors: | Tsai, C.J, Tani, K, Irie, K, Hiroaki, Y, Shimomura, T, Mcmillan, D.G, Cook, G.M, Schertler, G, Fujiyoshi, Y, Li, X.D. | | Deposit date: | 2013-03-28 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | ELECTRON CRYSTALLOGRAPHY (9 Å) | | Cite: | Two Alternative Conformations of a Voltage-Gated Sodium Channel.
J.Mol.Biol., 425, 2013
|
|
5TUG
 
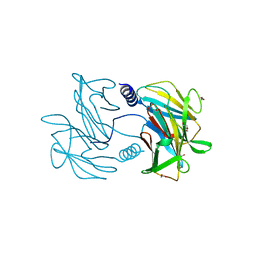 | |
5TUH
 
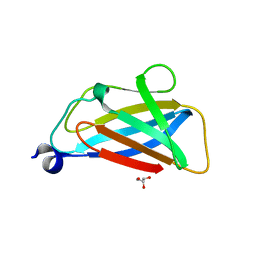 | |
4YDD
 
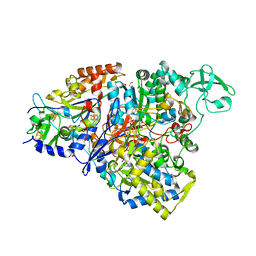 | | Crystal structure of the perchlorate reductase PcrAB from Azospira suillum PS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tsai, C.-L, Youngblut, M.D, Tainer, J.A. | | Deposit date: | 2015-02-21 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Perchlorate Reductase Is Distinguished by Active Site Aromatic Gate Residues.
J.Biol.Chem., 291, 2016
|
|
5CH7
 
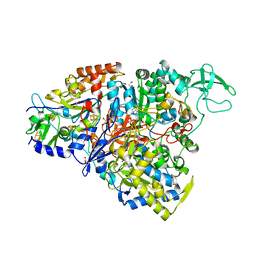 | | Crystal structure of the perchlorate reductase PcrAB - Phe164 gate switch intermediate - from Azospira suillum PS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ACETATE ION, ... | | Authors: | Tsai, C.-L, Tainer, J.A. | | Deposit date: | 2015-07-10 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Perchlorate Reductase Is Distinguished by Active Site Aromatic Gate Residues.
J.Biol.Chem., 291, 2016
|
|
5CHC
 
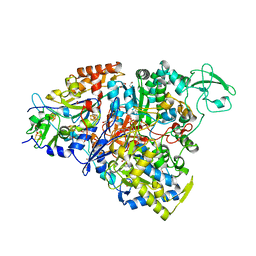 | | Crystal structure of the perchlorate reductase PcrAB - substrate analog SeO3 bound - from Azospira suillum PS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, BISELENITE ION, ... | | Authors: | Tsai, C.-L, Tainer, J.A. | | Deposit date: | 2015-07-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Perchlorate Reductase Is Distinguished by Active Site Aromatic Gate Residues.
J.Biol.Chem., 291, 2016
|
|
5E7O
 
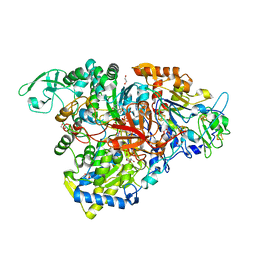 | | Crystal structure of the perchlorate reductase PcrAB mutant W461E of PcrA from Azospira suillum PS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DMSO reductase family type II enzyme, ... | | Authors: | Tsai, C.-L, Tainer, J.A. | | Deposit date: | 2015-10-12 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Perchlorate Reductase Is Distinguished by Active Site Aromatic Gate Residues.
J.Biol.Chem., 291, 2016
|
|
1OJ1
 
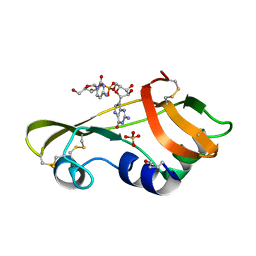 | | Nonproductive and Novel Binding Modes in Cytotoxic Ribonucleases from Rana catesbeiana of Two Crystal Structures Complexed with (2,5 CpG) and d(ApCpGpA) | | Descriptor: | CYTIDYL-2'-5'-PHOSPHO-GUANOSINE, RC-RNASE6 RIBONUCLEASE, SULFATE ION | | Authors: | Tsai, C.-J, Liu, J.-H, Liao, Y.-D, Chen, L.-y, Cheng, P.-T, Sun, Y.-J. | | Deposit date: | 2003-06-28 | | Release date: | 2004-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nonproductive and Novel Binding Modes in Cytotoxic Ribonucleases from Rana Catesbeiana of Two Crystal Structures Complexed with C(2,5 Cpg) and D(Apcpgpa)
To be Published
|
|
1OJ8
 
 | | Novel and retro Binding Modes in Cytotoxic Ribonucleases from Rana catesbeiana of Two Crystal Structures Complexed with d(ApCpGpA) and (2',5'CpG) | | Descriptor: | 5'-D(*AP*CP*GP*AP)-3', RC-RNASE6 RIBONUCLEASE, SULFATE ION | | Authors: | Tsai, C.-J, Liu, J.-H, Liao, Y.-D, Chen, L.-Y, Cheng, P.-T, Sun, Y.-J. | | Deposit date: | 2003-07-07 | | Release date: | 2004-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Retro and Novel Binding Modes in Cytotoxic Ribonucleases from Rana Catesbeiana of Two Crystal Structures Complexed with (2',5'Cpg) and D(Apcpgpa)
To be Published
|
|
4ZBH
 
 | |
2OZ0
 
 | | Mechanistic and Structural Studies of H373Q Flavocytochrome b2: Effects of Mutating the Active Site Base | | Descriptor: | Cytochrome b2, FLAVIN MONONUCLEOTIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tsai, C.-L, Gokulan, K, Sobrado, P, Sacchettini, J.C, Fitzpatrick, P.F. | | Deposit date: | 2007-02-23 | | Release date: | 2007-09-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and structural studies of H373Q flavocytochrome b2: effects of mutating the active site base.
Biochemistry, 46, 2007
|
|
3S5D
 
 | |
3S5E
 
 | | Crystal structure of human frataxin variant W155R, one of the Friedreich's ataxia point mutations | | Descriptor: | Frataxin, mitochondrial, MAGNESIUM ION | | Authors: | Tsai, C.-L, Bridwell-Rabb, J, Barondeau, D.P. | | Deposit date: | 2011-05-23 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Friedreich's Ataxia Variants I154F and W155R Diminish Frataxin-Based Activation of the Iron-Sulfur Cluster Assembly Complex.
Biochemistry, 50, 2011
|
|
3S5F
 
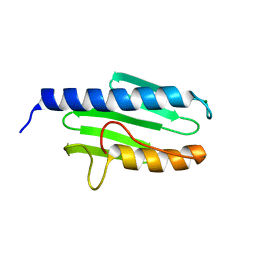 | |
7LWA
 
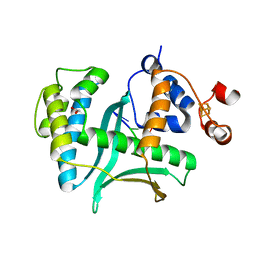 | |
7LW7
 
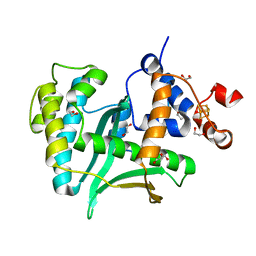 | | Human Exonuclease 5 crystal structure | | Descriptor: | 1,2-ETHANEDIOL, Exonuclease V, GLYCEROL, ... | | Authors: | Tsai, C.L, Tainer, J.A. | | Deposit date: | 2021-02-27 | | Release date: | 2021-07-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | EXO5-DNA structure and BLM interactions direct DNA resection critical for ATR-dependent replication restart.
Mol.Cell, 81, 2021
|
|
7LW9
 
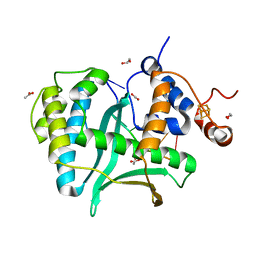 | | Human Exonuclease 5 crystal structure in complex with ssDNA, Sm, and Na | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*AP*TP*TP*GP*CP*TP*GP*AP*AP*GP*GP*G)-3'), ... | | Authors: | Tsai, C.L, Tainer, J.A. | | Deposit date: | 2021-02-28 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | EXO5-DNA structure and BLM interactions direct DNA resection critical for ATR-dependent replication restart.
Mol.Cell, 81, 2021
|
|
7LW8
 
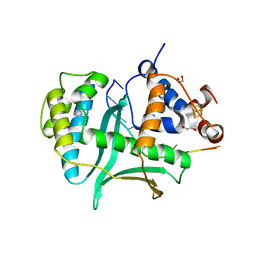 | | Human Exonuclease 5 crystal structure in complex with a ssDNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Exonuclease V, ... | | Authors: | Tsai, C.L, Tainer, J.A. | | Deposit date: | 2021-02-28 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | EXO5-DNA structure and BLM interactions direct DNA resection critical for ATR-dependent replication restart.
Mol.Cell, 81, 2021
|
|
3S4M
 
 | |
4P94
 
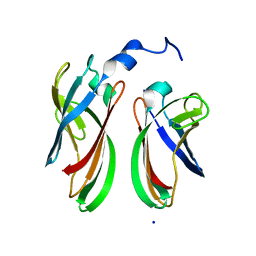 | | The crystal structure of the soluble domain of Sulfolobus acidocaldarius FlaF (residues 35-164) | | Descriptor: | Conserved flagellar protein F, SODIUM ION | | Authors: | Tsai, C.-L, Arvai, A.S, Ishida, J.P, Tainer, J.A. | | Deposit date: | 2014-04-02 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | FlaF Is a beta-Sandwich Protein that Anchors the Archaellum in the Archaeal Cell Envelope by Binding the S-Layer Protein.
Structure, 23, 2015
|
|
7O7F
 
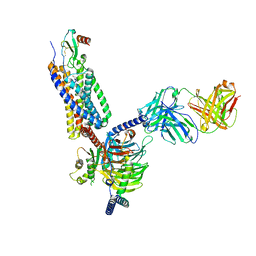 | | Structural basis of the activation of the CC chemokine receptor 5 by a chemokine agonist | | Descriptor: | C-C chemokine receptor type 5, C-C motif chemokine 5, Fab antibody fragment heavy chain, ... | | Authors: | Isaikina, P, Tsai, C.-J, Dietz, N.B, Pamula, F, Goldie, K.N, Schertler, G.F.X, Maier, T, Stahlberg, H, Deupi, X, Grzesiek, S. | | Deposit date: | 2021-04-13 | | Release date: | 2021-06-30 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis of the activation of the CC chemokine receptor 5 by a chemokine agonist.
Sci Adv, 7, 2021
|
|
