5M10
 
 | |
7UCA
 
 | | Horse liver alcohol dehydrogenase with NAD and trifluoroethanol at 65 K | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Alcohol dehydrogenase E chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), ... | | Authors: | Plapp, B.V, Gakhar, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Dependence of crystallographic atomic displacement factors on temperature (25-150 K) for complexes of horse liver alcohol dehydrogenases
Acta Crystallogr.,Sect.D, D78, 2022
|
|
8R3S
 
 | | Transketolase from Staphylococcus aureus in complex with thiamin pyrophosphate | | Descriptor: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, Transketolase | | Authors: | Ballut, L, Georges, N, Aghajari, N, Hecquet, L, Charmantray, F, Doumeche, B. | | Deposit date: | 2023-11-10 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Are Transketolases relevant Targets fighting Human Pathogens? A comparative Biochemical, Bioinformatic and Structural Study
To Be Published
|
|
7PR9
 
 | | Crystal structure of Burkholderia pseudomallei heparanase in complex with covalent inhibitor VL166 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-(2R,3S,5R,6R)-2,3,4,5,6-pentakis(oxidanyl)cyclohexane-1-carboxylic acid, Glyco_hydro_44 domain-containing protein | | Authors: | Wu, L, Armstrong, Z, Davies, G.J. | | Deposit date: | 2021-09-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Mechanism-based heparanase inhibitors reduce cancer metastasis in vivo.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3JBL
 
 | | Cryo-EM Structure of the Activated NAIP2/NLRC4 Inflammasome Reveals Nucleated Polymerization | | Descriptor: | NLR family CARD domain-containing protein 4 | | Authors: | Zhang, L, Chen, S, Ruan, J, Wu, J, Tong, A.B, Yin, Q, Li, Y, David, L, Lu, A, Wang, W.L, Marks, C, Ouyang, Q, Zhang, X, Mao, Y, Wu, H. | | Deposit date: | 2015-09-05 | | Release date: | 2015-10-21 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization.
Science, 350, 2015
|
|
7U3B
 
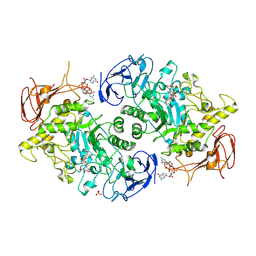 | | Structure of S. venezuelae GlgX bound to c-di-GMP and acarbose (pH 8.5) | | Descriptor: | 4-O-(4,6-dideoxy-4-{[(1S,2S,3S,4R,5S)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranosyl)-beta-D-glucopyranose, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Glycogen debranching enzyme GlgX, ... | | Authors: | Schumacher, M.A, Tschowri, N. | | Deposit date: | 2022-02-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Allosteric regulation of glycogen breakdown by the second messenger cyclic di-GMP.
Nat Commun, 13, 2022
|
|
2PB7
 
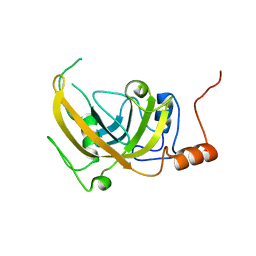 | |
3AI7
 
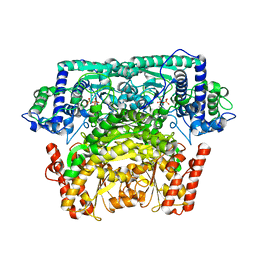 | | Crystal Structure of Bifidobacterium Longum Phosphoketolase | | Descriptor: | CALCIUM ION, THIAMINE DIPHOSPHATE, Xylulose-5-phosphate/fructose-6-phosphate phosphoketolase | | Authors: | Takahashi, K, Tagami, U, Shimba, N, Kashiwagi, T, Ishikawa, K, Suzuki, E. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Bifidobacterium Longum phosphoketolase; key enzyme for glucose metabolism in Bifidobacterium
Febs Lett., 584, 2010
|
|
7LUV
 
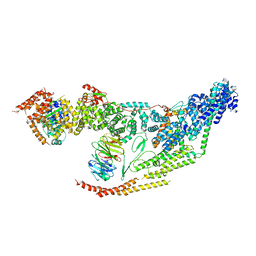 | | Cryo-EM structure of the yeast THO-Sub2 complex | | Descriptor: | ATP-dependent RNA helicase SUB2, THO complex subunit 2, THO complex subunit HPR1, ... | | Authors: | Xie, Y, Ren, Y. | | Deposit date: | 2021-02-23 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the yeast TREX complex and coordination with the SR-like protein Gbp2.
Elife, 10, 2021
|
|
2PBI
 
 | | The multifunctional nature of Gbeta5/RGS9 revealed from its crystal structure | | Descriptor: | GLYCEROL, Guanine nucleotide-binding protein subunit beta 5, Regulator of G-protein signaling 9 | | Authors: | Cheever, M.L, Snyder, J.T, Gershburg, S, Siderovski, D.P, Harden, T.K, Sondek, J. | | Deposit date: | 2007-03-28 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the multifunctional Gbeta5-RGS9 complex.
Nat.Struct.Mol.Biol., 15, 2008
|
|
7PTJ
 
 | |
3AKC
 
 | | Crystal structure of CMP kinase in complex with CDP and ADP from Thermus thermophilus HB8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CYTIDINE-5'-DIPHOSPHATE, Cytidylate kinase, ... | | Authors: | Mega, R, Nakagawa, N, Kuramitsu, S, Masui, R. | | Deposit date: | 2010-07-12 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of the tertiary complex of CMP kinase with a phosphoryl group acceptor and a donor from Thermus thermophilus HB8
To be Published
|
|
5M22
 
 | | Crystal structure of hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 | | Descriptor: | FE (III) ION, Hydroquinone dioxygenase large subunit, Hydroquinone dioxygenase small subunit | | Authors: | Ferraroni, M, Da Vela, S, Scozzafava, A, Kolvenbach, B, Corvini, P.F.X. | | Deposit date: | 2016-10-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structures of native hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 and of substrate and inhibitor complexes.
Biochim. Biophys. Acta, 1865, 2017
|
|
2GIN
 
 | |
7BK0
 
 | |
7BJ2
 
 | |
7U39
 
 | |
3AL3
 
 | | Crystal Structure of TopBP1 BRCT7/8-BACH1 peptide complex | | Descriptor: | DNA topoisomerase 2-binding protein 1, FORMIC ACID, Peptide of Fanconi anemia group J protein | | Authors: | Leung, C.C, Glover, J.N. | | Deposit date: | 2010-07-22 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular basis of BACH1/FANCJ recognition by TopBP1 in DNA replication checkpoint control
J.Biol.Chem., 286, 2011
|
|
2P6T
 
 | | CRYSTAL STRUCTURE OF TRANSCRIPTIONAL REGULATOR NMB0573 and L-LEUCINE COMPLEX FROM NEISSERIA MENINGITIDIS | | Descriptor: | CALCIUM ION, GLYCEROL, LEUCINE, ... | | Authors: | Ren, J, Sainsbury, S, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-03-19 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure and Transcriptional Analysis of a Global Regulator from Neisseria meningitidis.
J.Biol.Chem., 282, 2007
|
|
3K2H
 
 | | Co-crystal structure of dihydrofolate reductase/thymidylate synthase from Babesia bovis with dUMP, Pemetrexed and NADP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitor-bound complexes of dihydrofolate reductase-thymidylate synthase from Babesia bovis.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2OS7
 
 | | Caf1M periplasmic chaperone tetramer | | Descriptor: | Chaperone protein caf1M | | Authors: | Knight, S.D, Zavialov, A.Z. | | Deposit date: | 2007-02-05 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A novel self-capping mechanism controls aggregation of periplasmic chaperone Caf1M
MOL.MICROBIOL., 64, 2007
|
|
7Q46
 
 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of pericentriolar material 1 protein | | Descriptor: | CITRIC ACID, E3 ubiquitin-protein ligase HERC2, Pericentriolar material 1 protein | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.46002531 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of pericentriolar material 1 protein
To Be Published
|
|
7BHQ
 
 | |
5M59
 
 | | Crystal structure of Chaetomium thermophilum Brr2 helicase core in complex with Prp8 Jab1 domain | | Descriptor: | ACETATE ION, Pre-mRNA splicing helicase-like protein, Putative pre-mRNA splicing factor | | Authors: | Absmeier, E, Becke, C, Wollenhaupt, J, Santos, K.F, Wahl, M.C. | | Deposit date: | 2016-10-20 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Interplay of cis- and trans-regulatory mechanisms in the spliceosomal RNA helicase Brr2.
Cell Cycle, 16, 2017
|
|
3K38
 
 | |
