7AD1
 
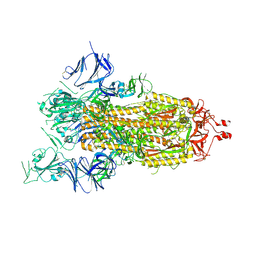 | | Cryo-EM structure of a prefusion stabilized SARS-CoV-2 Spike (D614N, R682S, R685G, A892P, A942P and V987P)(One up trimer) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein,Spike glycoprotein,Envelope glycoprotein,SARS-CoV-2 S protein | | Authors: | Rutten, L, Renault, L.L.R, Juraszek, J, Langedijk, J.P.M. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-04 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Stabilizing the closed SARS-CoV-2 spike trimer.
Nat Commun, 12, 2021
|
|
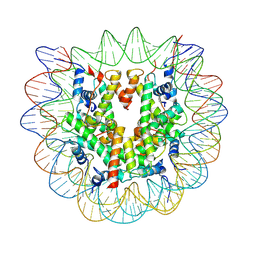
7U53
 
 | |
3JWU
 
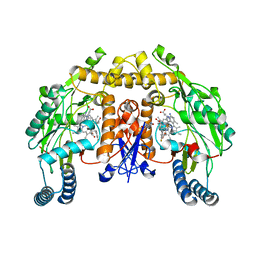 | | Structure of rat neuronal nitric oxide synthase R349A mutant heme domain in complex with N1-{(3'R,4'S)-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-fluorophenethyl)ethane-1,2-diamine tetrahydrochloride | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3R,4S)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}-N'-[2-(3-fluorophenyl)ethyl]ethane-1,2-diamine, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2009-09-18 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Unexpected binding modes of nitric oxide synthase inhibitors effective in the prevention of a cerebral palsy phenotype in an animal model.
J.Am.Chem.Soc., 132, 2010
|
|
3JX4
 
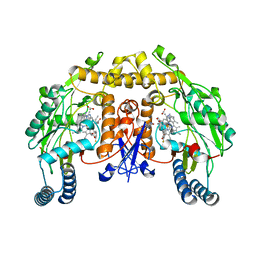 | | Structure of rat neuronal nitric oxide synthase D597N/M336V mutant heme domain in complex with N1-{(3'R,4'S)-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-fluorophenethyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3R,4S)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}-N'-[2-(3-fluorophenyl)ethyl]ethane-1,2-diamine, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2009-09-18 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Unexpected binding modes of nitric oxide synthase inhibitors effective in the prevention of a cerebral palsy phenotype in an animal model.
J.Am.Chem.Soc., 132, 2010
|
|
7TW8
 
 | |
3JXP
 
 | |
7U2X
 
 | | Cryo-EM structure of the pancreatic ATP-sensitive potassium channel in the presence of carbamazepine and ATP with Kir6.2-CTD in the down conformation | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shyng, S.L, Sung, M.W, Driggers, C.M. | | Deposit date: | 2022-02-25 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Ligand-mediated Structural Dynamics of a Mammalian Pancreatic K ATP Channel.
J.Mol.Biol., 434, 2022
|
|
5LOU
 
 | | human NUDT22 | | Descriptor: | Nucleoside diphosphate-linked moiety X motif 22, PHOSPHATE ION | | Authors: | Carter, M, Stenmark, P. | | Deposit date: | 2016-08-10 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human NUDT22 Is a UDP-Glucose/Galactose Hydrolase Exhibiting a Unique Structural Fold.
Structure, 26, 2018
|
|
3AMB
 
 | | Protein kinase A sixfold mutant model of Aurora B with inhibitor VX-680 | | Descriptor: | CYCLOPROPANECARBOXYLIC ACID {4-[4-(4-METHYL-PIPERAZIN-1-YL)-6-(5-METHYL-2H-PYRAZOL-3-YLAMINO)-PYRIMIDIN-2-YLSULFANYL]-PHENYL}-AMIDE, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Pflug, A, de Oliveira, T.M, Bossemeyer, D, Engh, R.A. | | Deposit date: | 2010-08-18 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mutants of protein kinase A that mimic the ATP-binding site of Aurora kinase
Biochem.J., 440, 2011
|
|
2JGL
 
 | | Crystal structure of mouse acetylcholinesterase inhibited by aged VX and sarin | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Hornberg, A, Tunemalm, A.-K, Ekstrom, F. | | Deposit date: | 2007-02-13 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Acetylcholinesterase in Complex with Organophosphorus Compounds Suggest that the Acyl Pocket Modulates the Aging Reaction by Precluding the Formation of the Trigonal Bipyramidal Transition State.
Biochemistry, 46, 2007
|
|
7LMW
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with 3-(3-((4-(4-carboxyphenoxy)benzyl)oxy)phenyl)-1H-indole-2-carboxylic acid | | Descriptor: | 7-methyl-3-(1~{H}-pyrazol-4-yl)-1~{H}-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2021-02-06 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
3AOG
 
 | |
2JBM
 
 | | QPRTASE STRUCTURE FROM HUMAN | | Descriptor: | NICOTINATE-NUCLEOTIDE PYROPHOSPHORYLASE, S,R MESO-TARTARIC ACID | | Authors: | Liu, H, Naismith, J.H. | | Deposit date: | 2006-12-08 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Characterization of Quinolinate Phosphoribosyltransferase (Hqprtase) from Homo Sapiens.
J.Mol.Biol., 373, 2007
|
|
5LTQ
 
 | |
3JAV
 
 | | Structure of full-length IP3R1 channel in the apo-state determined by single particle cryo-EM | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Fan, G, Baker, M.L, Wang, Z, Baker, M.R, Sinyagovskiy, P.A, Chiu, W, Ludtke, S.J, Serysheva, I.I. | | Deposit date: | 2015-06-30 | | Release date: | 2015-10-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Gating machinery of InsP3R channels revealed by electron cryomicroscopy.
Nature, 527, 2015
|
|
2N6P
 
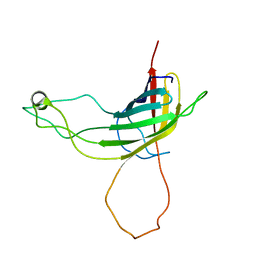 | | Solution NMR structure of Outer Membrane Protein G P92A mutant from Pseudomonas aeruginosa | | Descriptor: | Outer membrane protein OprG | | Authors: | Kucharska, I, Seelheim, P, Edrington, T.C, Liang, B, Tamm, L.K. | | Deposit date: | 2015-08-27 | | Release date: | 2015-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | OprG Harnesses the Dynamics of its Extracellular Loops to Transport Small Amino Acids across the Outer Membrane of Pseudomonas aeruginosa.
Structure, 23, 2015
|
|
3J92
 
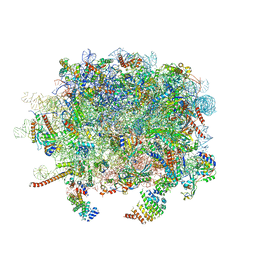 | | Structure and assembly pathway of the ribosome quality control complex | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Shao, S, Brown, A, Santhanam, B, Hegde, R.S. | | Deposit date: | 2014-12-02 | | Release date: | 2015-01-21 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and Assembly Pathway of the Ribosome Quality Control Complex.
Mol.Cell, 57, 2015
|
|
7PMZ
 
 | | Crystal structure of Streptomyces coelicolor guaB (IMP dehydrogenase) bound to ATP and ppGpp at 2.0 A resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5',3'-TETRAPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Fernandez-Justel, D, Revuelta, J.L, Buey, R.M. | | Deposit date: | 2021-09-04 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Diversity of mechanisms to control bacterial GTP homeostasis by the mutually exclusive binding of adenine and guanine nucleotides to IMP dehydrogenase.
Protein Sci., 31, 2022
|
|
5LVL
 
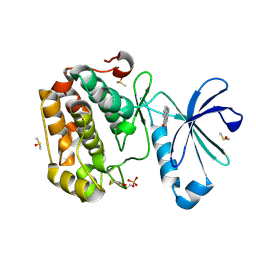 | | Human PDK1 Kinase Domain in Complex with Compound PS653 Bound to the ATP-Binding Site | | Descriptor: | 2,6-DIHYDROANTHRA/1,9-CD/PYRAZOL-6-ONE, 3-phosphoinositide-dependent protein kinase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Schulze, J.O, Saladino, G, Busschots, K, Neimanis, S, Suess, E, Odadzic, D, Zeuzem, S, Hindie, V, Herbrand, A.K, Lisa, M.N, Alzari, P.M, Gervasio, F.L, Biondi, R.M. | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Bidirectional Allosteric Communication between the ATP-Binding Site and the Regulatory PIF Pocket in PDK1 Protein Kinase.
Cell Chem Biol, 23, 2016
|
|
3AF7
 
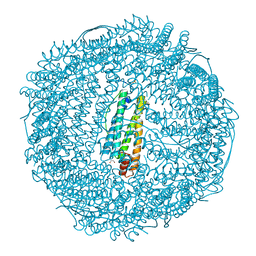 | | Crystal Structure of 25Pd(allyl)/apo-Fr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Abe, S, Hikage, T, Watanabe, Y, Kitagawa, S, Ueno, T. | | Deposit date: | 2010-02-24 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Mechanism of Accumulation and Incorporation of Organometallic Pd Complexes into the Protein Nanocage of apo-Ferritin.
Inorg.Chem., 49, 2010
|
|
3JCA
 
 | | Core model of the Mouse Mammary Tumor Virus intasome | | Descriptor: | 5'-D(*AP*AP*TP*GP*CP*CP*GP*CP*AP*GP*TP*CP*GP*GP*CP*CP*GP*AP*CP*CP*TP*G)-3', 5'-D(*CP*AP*GP*GP*TP*CP*GP*GP*CP*CP*GP*AP*CP*TP*GP*CP*GP*GP*CP*A)-3', Integrase, ... | | Authors: | Lyumkis, D.L, Ballandras-Colas, A, Brown, M, Cook, N.J, Dewdney, T.G, Demeler, B, Cherepanov, P, Engelman, A.N. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function.
Nature, 530, 2016
|
|
7PVH
 
 | |
7LVI
 
 | | CRYSTAL STRUCTURE OF AP2 ASSOCIATED KINASE 1 ISOFORM 1 COMPLEXED WITH LIGAND (2R)-2-AMINO-N-[3-METHOXY-4- (1,3-OXAZOL-5-YL)PHENYL]-4-METHYLPENTANAMIDE | | Descriptor: | (2R,3R)-N-[3-methoxy-4-(1,3-oxazol-5-yl)phenyl]-3-(propan-2-yl)piperidine-2-carboxamide, 5-[(4-aminopiperidin-1-yl)methyl]-N-{3-[5-(propan-2-yl)-1,3,4-thiadiazol-2-yl]phenyl}pyrrolo[2,1-f][1,2,4]triazin-4-amine, AP2-associated protein kinase 1, ... | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery, Structure-Activity Relationships, and In Vivo Evaluation of Novel Aryl Amides as Brain Penetrant Adaptor Protein 2-Associated Kinase 1 (AAK1) Inhibitors for the Treatment of Neuropathic Pain.
J.Med.Chem., 64, 2021
|
|
2NA0
 
 | |
5LWD
 
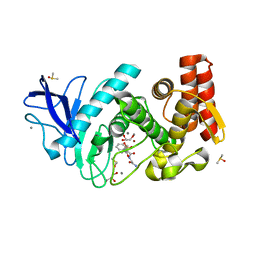 | | Thermolysin in complex with inhibitor (JC96) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-09-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Elucidating the Origin of Long Residence Time Binding for Inhibitors of the Metalloprotease Thermolysin.
ACS Chem. Biol., 12, 2017
|
|
