3HOZ
 
 | | Complete RNA polymerase II elongation complex IV with a T-U mismatch and a frayed RNA 3'-guanine | | Descriptor: | 5'-D(*AP*CP*TP*AP*CP*TP*TP*GP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*C*AP*AP*GP*TP*AP*GP*TP*TP*CP*TP*GP*CP*CP*(BRU)P*GP*GP*TP*CP*AP*TP*T)-3', 5'-R(*UP*GP*CP*AP*UP*UP*U*CP*AP*AP*CP*CP*AP*GP*GP*CP*UP*G)-3', ... | | Authors: | Sydow, J.F, Brueckner, F, Cheung, A.C.M, Damsma, G.E, Dengl, S, Lehmann, E, Vassylyev, D, Cramer, P. | | Deposit date: | 2009-06-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA.
Mol.Cell, 34, 2009
|
|
3I4N
 
 | | 8-oxoguanine containing RNA polymerase II elongation complex E | | Descriptor: | DNA (5'-D(*AP*G*CP*TP*CP*AP*AP*GP*TP*AP*CP*TP*TP*AP*(8OG)P*GP*CP*CP*(BRU)P*GP*GP*TP*CP*AP*TP*T)-3'), DNA (5'-D(*AP*GP*TP*AP*CP*TP*TP*GP*AP*GP*CP*T)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Damsma, G.E, Cramer, P. | | Deposit date: | 2009-07-02 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Molecular basis of transcriptional mutagenesis at 8-oxoguanine
J.Biol.Chem., 284, 2009
|
|
3J0K
 
 | | Orientation of RNA polymerase II within the human VP16-Mediator-pol II-TFIIF assembly | | Descriptor: | DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 140 kDa polypeptide, DNA-directed RNA polymerase II 19 kDa polypeptide, ... | | Authors: | Bernecky, C, Grob, P, Ebmeier, C.C, Nogales, E, Taatjes, D.J. | | Deposit date: | 2011-10-04 | | Release date: | 2011-10-19 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (36 Å) | | Cite: | Molecular architecture of the human Mediator-RNA polymerase II-TFIIF assembly.
Plos Biol., 9, 2011
|
|
3H3V
 
 | | Yeast RNAP II containing poly(A)-signal sequence in the active site | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP*GP*CP*TP*GP*CP*TP*TP*TP*AP*TP*TP*GP*CP*AP*TP*T)-3', 5'-D(*CP*AP*GP*CP*TP*AP*CP*TP*TP*GP*AP*GP*CP*T)-3', 5'-R(*UP*GP*CP*AP*UP*UP*UP*CP*GP*CP*AP*AP*UP*AP*AP*A)-3', ... | | Authors: | Dengl, S, Cramer, P. | | Deposit date: | 2009-04-17 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Torpedo Nuclease Rat1 Is Insufficient to Terminate RNA Polymerase II in Vitro
J.Biol.Chem., 284, 2009
|
|
3HOV
 
 | | Complete RNA polymerase II elongation complex II | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*A*GP*TP*AP*GP*TP*TP*AP*TP*GP*CP*CP*(BRU)P*GP*GP*TP*CP*AP*TP*T)-3', 5'-D(*T*AP*CP*TP*AP*CP*TP*TP*GP*AP*GP*CP*T)-3', 5'-R(*UP*GP*CP*AP*UP*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*CP*A)-3', ... | | Authors: | Sydow, J.F, Brueckner, F, Cheung, A.C.M, Damsma, G.E, Dengl, S, Lehmann, E, Vassylyev, D, Cramer, P. | | Deposit date: | 2009-06-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA.
Mol.Cell, 34, 2009
|
|
3J81
 
 | | CryoEM structure of a partial yeast 48S preinitiation complex | | Descriptor: | 18S rRNA, MAGNESIUM ION, METHIONINE, ... | | Authors: | Hussain, T, Llacer, J.L, Fernandez, I.S, Savva, C.G, Ramakrishnan, V. | | Deposit date: | 2014-08-29 | | Release date: | 2014-11-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural changes enable start codon recognition by the eukaryotic translation initiation complex.
Cell(Cambridge,Mass.), 159, 2014
|
|
3I4M
 
 | | 8-oxoguanine containing RNA polymerase II elongation complex D | | Descriptor: | DNA (5'-D(*AP*G*CP*TP*CP*AP*AP*GP*TP*AP*CP*TP*TP*AP*(8OG)P*GP*CP*CP*(BRU)P*GP*GP*TP*CP*AP*TP*T)-3'), DNA (5'-D(*AP*GP*TP*AP*CP*TP*TP*GP*AP*GP*CP*T)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Damsma, G.E, Cramer, P. | | Deposit date: | 2009-07-02 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Molecular basis of transcriptional mutagenesis at 8-oxoguanine
J.Biol.Chem., 284, 2009
|
|
3JAP
 
 | | Structure of a partial yeast 48S preinitiation complex in closed conformation | | Descriptor: | 18S rRNA, MAGNESIUM ION, METHIONINE, ... | | Authors: | Llacer, J.L, Hussain, T, Ramakrishnan, V. | | Deposit date: | 2015-06-18 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Conformational Differences between Open and Closed States of the Eukaryotic Translation Initiation Complex.
Mol.Cell, 59, 2015
|
|
3K1F
 
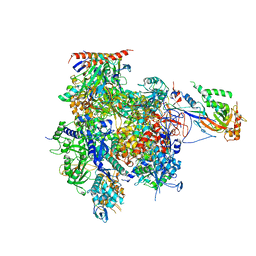 | | Crystal structure of RNA Polymerase II in complex with TFIIB | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Kostrewa, D, Zeller, M.E, Armache, K.-J, Seizl, M, Leike, K, Thomm, M, Cramer, P. | | Deposit date: | 2009-09-27 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | RNA polymerase II-TFIIB structure and mechanism of transcription initiation.
Nature, 462, 2009
|
|
3J1N
 
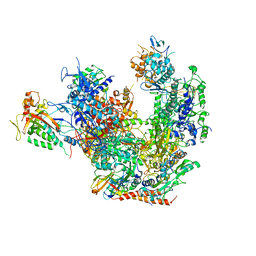 | |
8S5N
 
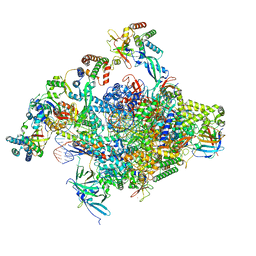 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 12 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-24 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8S55
 
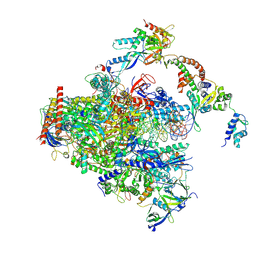 | | RNA polymerase II early elongation complex bound to TFIIE and TFIIF - state a (composite structure) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8S51
 
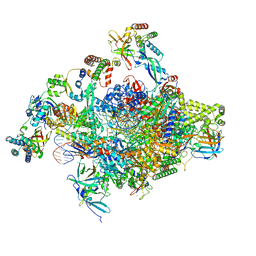 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 8 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8S52
 
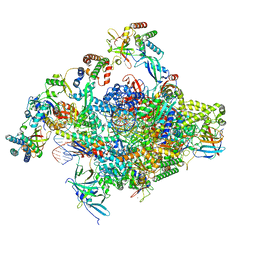 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 10 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8S54
 
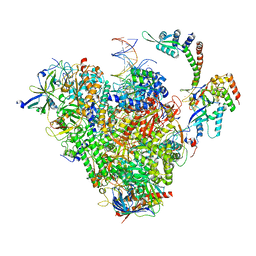 | | RNA polymerase II early elongation complex bound to TFIIE and TFIIF - state b (composite structure) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabber, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8UI0
 
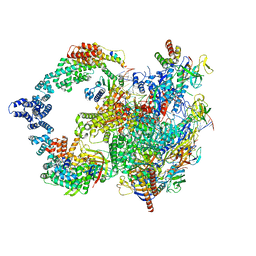 | |
8UHG
 
 | |
8UIS
 
 | | Structure of transcription complex Pol II-DSIF-NELF-TFIIS | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB3, DNA-directed RNA polymerase II subunit RPB7, ... | | Authors: | Su, B.G, Vos, S.M. | | Deposit date: | 2023-10-10 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Distinct negative elongation factor conformations regulate RNA polymerase II promoter-proximal pausing.
Mol.Cell, 84, 2024
|
|
8UHA
 
 | |
8UHD
 
 | | Structure of paused transcription complex Pol II-DSIF-NELF - post-translocated | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB3, DNA-directed RNA polymerase II subunit RPB7, ... | | Authors: | Su, B.G, Vos, S.M. | | Deposit date: | 2023-10-08 | | Release date: | 2024-03-20 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Distinct negative elongation factor conformations regulate RNA polymerase II promoter-proximal pausing.
Mol.Cell, 84, 2024
|
|
8TVP
 
 | | Cryo-EM structure of CPD-stalled Pol II in complex with Rad26 (open state) | | Descriptor: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVV
 
 | | Cryo-EM structure of backtracked Pol II | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVY
 
 | | Cryo-EM structure of CPD lesion containing RNA Polymerase II elongation complex with Rad26 and Elf1 (closed state) | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVX
 
 | | Cryo-EM structure of CPD-stalled Pol II (Conformation 2) | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVW
 
 | | Cryo-EM structure of CPD-stalled Pol II (conformation 1) | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
