4QOO
 
 | | Structure of Bacillus pumilus catalase with resorcinol bound. | | Descriptor: | CHLORIDE ION, Catalase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
3VT4
 
 | | Crystal structures of rat VDR-LBD with R270L mutation | | Descriptor: | (1R,2Z,3R,5E,7E)-17-{(1S)-1-[(2-ethyl-2-hydroxybutyl)sulfanyl]ethyl}-2-(2-hydroxyethylidene)-9,10-secoestra-5,7,16-triene-1,3-diol, COACTIVATOR PEPTIDE DRIP, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Shimizu, M, Ikura, T, Ito, N. | | Deposit date: | 2012-05-19 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of hereditary vitamin D-resistant rickets-associated vitamin D receptor mutants R270L and W282R bound to 1,25-dihydroxyvitamin D3 and synthetic ligands.
J.Med.Chem., 56, 2013
|
|
4QN8
 
 | | The crystal structure of an effector protein VipE from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | BETA-MERCAPTOETHANOL, VipE | | Authors: | Tan, K, Xu, X, Cui, H, Liu, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-16 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | The crystal structure of an effector protein VipE from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
To be Published
|
|
2NZI
 
 | |
1CLX
 
 | | CATALYTIC CORE OF XYLANASE A | | Descriptor: | CALCIUM ION, XYLANASE A | | Authors: | Harris, G.W, Jenkins, J.A, Connerton, I, Pickersgill, R.W. | | Deposit date: | 1995-08-31 | | Release date: | 1996-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined crystal structure of the catalytic domain of xylanase A from Pseudomonas fluorescens at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2AR0
 
 | |
3C34
 
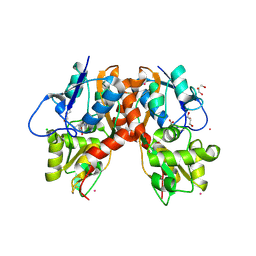 | |
4R02
 
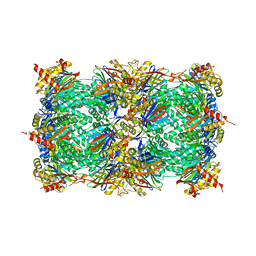 | | yCP in complex with BSc4999 (alpha-Keto Phenylamide) | | Descriptor: | MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-{(2S,3S)-1-[(2,4-dimethylphenyl)amino]-2-hydroxy-5-methyl-1-oxohexan-3-yl}-L-leucinamide, Probable proteasome subunit alpha type-7, ... | | Authors: | Voss, C, Scholz, C, Knorr, S, Beck, P, Stein, M, Zall, A, Kuckelkorn, U, Kloetzel, P.-M, Groll, M, Hamacher, K, Schmidt, B. | | Deposit date: | 2014-07-29 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | alpha-Keto Phenylamides as P1'-Extended Proteasome Inhibitors.
Chemmedchem, 9, 2014
|
|
2XH2
 
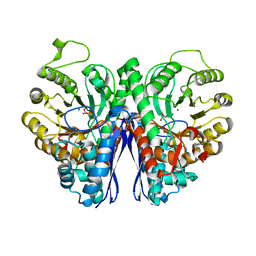 | |
3R6J
 
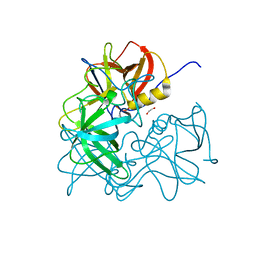 | | Crystal Structure of the Capsid P Domain from Norwalk Virus Strain Hiroshima/1999 | | Descriptor: | 1,2-ETHANEDIOL, VP1 protein | | Authors: | Hansman, G.S, Biertumpfel, C, McLellan, J.S, Georgiev, I, Chen, L, Zhou, T, Katayama, K, Kwong, P.D. | | Deposit date: | 2011-03-21 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of GII.10 and GII.12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability.
J.Virol., 85, 2011
|
|
3CU2
 
 | |
3O2L
 
 | |
2XD9
 
 | | STRUCTURE OF HELICOBACTER PYLORI TYPE II DEHYDROQUINASE IN COMPLEX WITH INHIBITOR COMPOUND (4R,6R,7S)-4,6,7-Trihydroxy-2-((E)-prop-1- enyl)-4,5,6,7-tetrahydrobenzo(b)thiophene-4-carboxylic acid | | Descriptor: | (4R,6R,7S)-4,6,7-TRIHYDROXY-2-[(1E)-PROP-1-EN-1-YL]-4,5,6,7-TETRAHYDRO-1-BENZOTHIOPHENE-4-CARBOXYLIC ACID, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Paz, S, Tizon, L, Otero, J.M, Llamas-Saiz, A.L, Fox, G.C, van Raaij, M.J, Lamb, H, Hawkins, A.R, Lapthorn, A.J, Castedo, L, Gonzalez-Bello, C. | | Deposit date: | 2010-04-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tetrahydrobenzothiophene derivatives: conformationally restricted inhibitors of type II dehydroquinase.
ChemMedChem, 6, 2011
|
|
3GDZ
 
 | | Crystal structure of arginyl-tRNA synthetase from Klebsiella pneumoniae subsp. pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, Arginyl-tRNA synthetase | | Authors: | Chang, C, Wu, R, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of arginyl-tRNA synthetase from Klebsiella pneumoniae subsp. pneumoniae
To be Published
|
|
3RB3
 
 | |
4F76
 
 | |
4MDB
 
 | | Structure of Mos1 transposase catalytic domain and Raltegravir with Mg | | Descriptor: | MAGNESIUM ION, Mariner Mos1 transposase, N-(4-fluorobenzyl)-5-hydroxy-1-methyl-2-(1-methyl-1-{[(5-methyl-1,3,4-oxadiazol-2-yl)carbonyl]amino}ethyl)-6-oxo-1,6-di hydropyrimidine-4-carboxamide | | Authors: | Richardson, J.M. | | Deposit date: | 2013-08-22 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Mos1 Transposase Inhibition by the Anti-retroviral Drug Raltegravir.
Acs Chem.Biol., 9, 2014
|
|
2R8Q
 
 | | Structure of LmjPDEB1 in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, Class I phosphodiesterase PDEB1, MAGNESIUM ION, ... | | Authors: | Wang, H, Yan, Z, Geng, J, Kunz, S, Seebeck, T, Ke, H. | | Deposit date: | 2007-09-11 | | Release date: | 2007-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the Leishmania major phosphodiesterase LmjPDEB1 and insight into the design of the parasite-selective inhibitors.
Mol.Microbiol., 66, 2007
|
|
3NPD
 
 | |
3DJ7
 
 | | Crystal structure of the mouse Aurora-A catalytic domain (Asn186->Gly, Lys240->Arg, Met302->Leu) in complex with Compound 130. | | Descriptor: | 1-(5-{2-[(6-amino-5-bromopyrimidin-4-yl)amino]ethyl}-1,3-thiazol-2-yl)-3-[3-(trifluoromethyl)phenyl]urea, serine/threonine kinase 6 | | Authors: | Elling, R.A, Yang, W, Erlanson, D.A, Tangonan, B.T, Hansen, S.K, Romanowski, M.J. | | Deposit date: | 2008-06-22 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | New fragment-based drug discovery
To be Published
|
|
1TC1
 
 | | A 1.4 ANGSTROM CRYSTAL STRUCTURE FOR THE HYPOXANTHINE PHOSPHORIBOSYLTRANSFERASE OF TRYPANOSOMA CRUZI | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FORMYCIN B, PROTEIN (HYPOXANTHINE PHOSPHORIBOSYLTRANSFERASE) | | Authors: | Focia, P.J, Craig III, S.P, Nieves-Alicea, R, Fletterick, R.J, Eakin, A.E. | | Deposit date: | 1998-09-30 | | Release date: | 1999-10-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A 1.4 A crystal structure for the hypoxanthine phosphoribosyltransferase of Trypanosoma cruzi.
Biochemistry, 37, 1998
|
|
3DR4
 
 | | GDP-perosamine synthase K186A mutant from Caulobacter crescentus with bound sugar ligand | | Descriptor: | 1,2-ETHANEDIOL, Putative perosamine synthetase, [(2R,3S,4R,5R)-5-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S,5S,6R)-3,4-dihydroxy-5-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-6-methyltetrahydro-2H-pyran-2-yl dihydrogen diphosphate | | Authors: | Holden, H.M, Cook, P.D, Carney, A.E. | | Deposit date: | 2008-07-10 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Accommodation of GDP-linked sugars in the active site of GDP-perosamine synthase
Biochemistry, 47, 2008
|
|
4J0P
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH 5-Cyano-pyridine-2-carboxylic acid [3-((S)-2-amino-4-methyl-5,6-dihydro-4H-[1,3]oxazin-4-yl)-4-fluoro-phenyl]-amide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, N-{3-[(4S)-2-amino-4-methyl-5,6-dihydro-4H-1,3-oxazin-4-yl]-4-fluorophenyl}-5-cyanopyridine-2-carboxamide, ... | | Authors: | Kuglstatter, A, Stihle, M. | | Deposit date: | 2013-01-31 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | beta-Secretase (BACE1) inhibitors with high in vivo efficacy suitable for clinical evaluation in Alzheimer's disease.
J.Med.Chem., 56, 2013
|
|
4R5A
 
 | |
4J1H
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH 5-Cyano-pyridine-2-carboxylic acid [3-((4S,6R)-2-amino-4-methyl-6-trifluoromethyl-5,6-dihydro-4H-[1,3]oxazin-4-yl)-4-fluoro-phenyl]-amide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, N-{3-[(4S,6R)-2-amino-4-methyl-6-(trifluoromethyl)-5,6-dihydro-4H-1,3-oxazin-4-yl]-4-fluorophenyl}-5-cyanopyridine-2-carboxamide, ... | | Authors: | Kuglstatter, A, Stihle, M. | | Deposit date: | 2013-02-01 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | beta-Secretase (BACE1) Inhibitors with High In Vivo Efficacy Suitable for Clinical Evaluation in Alzheimer s Disease
J.Med.Chem., 56, 2013
|
|
