5NJZ
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 1g | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-(piperidin-4-ylcarbamoyl)phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NK1
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 1k | | Descriptor: | 2-[[3-[(4-azanyl-6-methyl-1,3,5-triazin-2-yl)carbamoyl]phenyl]amino]-~{N}-(2-chloranyl-6-methyl-phenyl)-1,3-thiazole-5-carboxamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NK6
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2d | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-[(4-oxidanylcyclohexyl)carbamoyl]phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.267 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5MZZ
 
 | | Crystal structure of the decarboxylase AibA/AibB in complex with 3-methylglutaconate | | Descriptor: | 3-methylpent-2-enedioic acid, ACETATE ION, GLYCEROL, ... | | Authors: | Bock, T, Luxenburger, E, Hoffmann, J, Schuetza, V, Feiler, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AibA/AibB Induces an Intramolecular Decarboxylation in Isovalerate Biosynthesis by Myxococcus xanthus.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5NJF
 
 | | E. coli Microcin-processing metalloprotease TldD/E (TldD H262A mutant) with pentapeptide bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ALA-ALA-ALA-ALA-ALA, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5NK2
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2b | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-[[(3~{R},4~{R})-3-fluoranylpiperidin-4-yl]carbamoyl]phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NKA
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2g | | Descriptor: | 1,2-ETHANEDIOL, 4-[[3-[[5-[(2-chloranyl-6-methyl-phenyl)carbamoyl]-1,3-thiazol-2-yl]amino]phenyl]carbonylamino]cyclohexane-1-carboxylic acid, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.377 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NKJ
 
 | | X-ray structure of the N239C mutant of GLIC | | Descriptor: | Proton-gated ion channel | | Authors: | Hu, H, Fourati, Z, Delarue, M. | | Deposit date: | 2017-03-31 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.74 Å) | | Cite: | Structural Basis for a Bimodal Allosteric Mechanism of General Anesthetic Modulation in Pentameric Ligand-Gated Ion Channels.
Cell Rep, 23, 2018
|
|
5N1U
 
 | |
5N2H
 
 | | Structure of the E9 DNA polymerase exonuclease deficient mutant (D166A+E168A) from vaccinia virus | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tarbouriech, N, Burmeister, W.P, Iseni, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding.
Nat Commun, 8, 2017
|
|
5N2W
 
 | | WT-Parkin and pUB complex | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin,E3 ubiquitin-protein ligase parkin, Polyubiquitin-B, ... | | Authors: | Kumar, A, Chaugule, V.K, Johnson, C, Toth, R, Sundaramoorthy, R, Knebel, A, Walden, H. | | Deposit date: | 2017-02-08 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Parkin-phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5NN6
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with N-hydroxyethyl-1-deoxynojirimycin | | Descriptor: | (2R,3R,4R,5S)-1-(2-hydroxyethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roig-Zamboni, V, Cobucci-Ponzano, B, Iacono, R, Ferrara, M.C, Germany, S, Parenti, G, Bourne, Y, Moracci, M. | | Deposit date: | 2017-04-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human lysosomal acid alpha-glucosidase-a guide for the treatment of Pompe disease.
Nat Commun, 8, 2017
|
|
5NNJ
 
 | | Dimer structure of Sortilin ectodomain crystal form 3, 4.0 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Sortilin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Leloup, N.O.L, Janssen, B.J.C. | | Deposit date: | 2017-04-09 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun, 8, 2017
|
|
5N5E
 
 | |
5O5U
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a urea based inhibitor PSMA 1027 | | Descriptor: | (2~{S})-2-[[(2~{S})-6-[[(2~{S})-2-[[4-[[[(2~{S})-2-[[(2~{R})-2-[(6-fluoranylpyridin-3-yl)carbonylamino]-3-sulfo-propanoyl]amino]-3-sulfo-propanoyl]amino]methyl]phenyl]carbonylamino]-3-naphthalen-2-yl-propanoyl]amino]-1-oxidanyl-1-oxidanylidene-hexan-2-yl]carbamoylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Novakova, Z, Barinka, C, Motlova, L. | | Deposit date: | 2017-06-02 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a urea based inhibitor PSMA 1027
To Be Published
|
|
5O8F
 
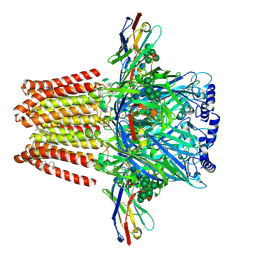 | | Structure of a chimaeric beta3-alpha5 GABAA receptor in complex with nanobody Nb25 and pregnanolone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit beta-3,Gamma-aminobutyric acid receptor subunit alpha-5,Gamma-aminobutyric acid receptor subunit alpha-5, Nanobody Nb25, ... | | Authors: | Miller, P.S, Scott, S, Masiulis, S, De Colibus, L, Pardon, E, Steyaert, J, Aricescu, A.R. | | Deposit date: | 2017-06-13 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for GABAA receptor potentiation by neurosteroids.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5O7D
 
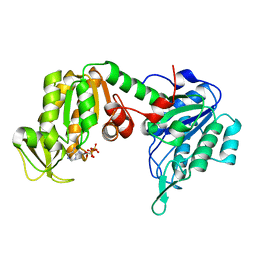 | | The X-ray structure of human R38M phosphoglycerate kinase 1 mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphoglycerate kinase 1 | | Authors: | Ilari, A, Fiorillo, A, Cipollone, A, Petrosino, M. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The phosphoglycerate kinase 1 variants found in carcinoma cells display different catalytic activity and conformational stability compared to the native enzyme.
PLoS ONE, 13, 2018
|
|
5O7I
 
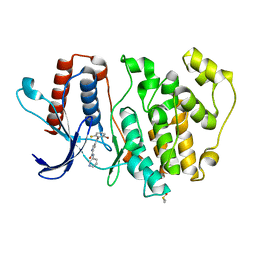 | | ERK5 in complex with a pyrrole inhibitor | | Descriptor: | 4-(2-bromanyl-6-fluoranyl-phenyl)carbonyl-~{N}-pyridin-3-yl-1~{H}-pyrrole-2-carboxamide, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 7 | | Authors: | Tucker, J.A, Heptinstall, A, Myers, S. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Identification of a novel orally bioavailable ERK5 inhibitor with selectivity over p38 alpha and BRD4.
Eur.J.Med.Chem., 178, 2019
|
|
5O7P
 
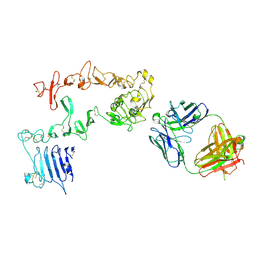 | | HER3 in complex with Fab MF3178 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MF3178 FAB heavy chain, MF3178 FAB light chain, ... | | Authors: | De Nardis, C, Gros, P. | | Deposit date: | 2017-06-09 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Unbiased Combinatorial Screening Identifies a Bispecific IgG1 that Potently Inhibits HER3 Signaling via HER2-Guided Ligand Blockade.
Cancer Cell, 33, 2018
|
|
5NVW
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-(cyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 6) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-(cyclopropylcarbonylamino)-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, Ciulli, A. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298).
J. Med. Chem., 61, 2018
|
|
5O97
 
 | |
5NYZ
 
 | | Twist and induce: Dissecting the link between the enzymatic activity and the SaPI inducing capacity of the phage 80 dUTPase. D95E mutant from dUTPase 80alpha phage. | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPase, MAGNESIUM ION | | Authors: | Alite, C, Humphrey, S, Donderis, J, Maiques, E, Ciges-Tomas, J.R, Penades, J.R, Marina, A. | | Deposit date: | 2017-05-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dissecting the link between the enzymatic activity and the SaPI inducing capacity of the phage 80 alpha dUTPase.
Sci Rep, 7, 2017
|
|
5OAZ
 
 | |
5NSG
 
 | | Fc DEDE homodimer variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Putative uncharacterized protein DKFZp686C11235, ... | | Authors: | De Nardis, C, Hendriks, L.J.A, Poirier, E, Arvinte, T, Gros, P, Bakker, A.B.H, de Kruif, J. | | Deposit date: | 2017-04-26 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A new approach for generating bispecific antibodies based on a common light chain format and the stable architecture of human immunoglobulin G1.
J. Biol. Chem., 292, 2017
|
|
5NSK
 
 | | apo Structure of Leucyl aminopeptidase from Trypanosoma brucei | | Descriptor: | Aminopeptidase, putative | | Authors: | Timm, J, Wilson, K.S. | | Deposit date: | 2017-04-26 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Characterization of Acidic M17 Leucine Aminopeptidases from the TriTryps and Evaluation of Their Role in Nutrient Starvation inTrypanosoma brucei.
mSphere, 2, 2018
|
|
