8IF2
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Omicron BQ.1.1 variant spike protein in complex with its receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Kimura, K, Suzuki, T, Hashiguchi, T. | | Deposit date: | 2023-02-17 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Convergent evolution of SARS-CoV-2 Omicron subvariants leading to the emergence of BQ.1.1 variant.
Nat Commun, 14, 2023
|
|
7UJ1
 
 | | Crystal structure of PSF-RNA complex | | Descriptor: | RNA (30-MER), Splicing factor, proline- and glutamine-rich | | Authors: | Sachpatzidis, A, Wang, J, Konigsberg, W.H. | | Deposit date: | 2022-03-30 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Insight into the Tumor Suppression Mechanism from the Structure of Human Polypyrimidine Splicing Factor (PSF/SFPQ) Complexed with a 30mer RNA from Murine Virus-like 30S Transcript-1.
Biochemistry, 61, 2022
|
|
4M3G
 
 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | 4-(2-methyl-1,3-thiazol-4-yl)-N-(3,3,3-trifluoropropyl)benzenesulfonamide, HTH-type transcriptional regulator EthR | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
4P4K
 
 | | Structural Basis of Chronic Beryllium Disease: Bridging the Gap Between allergic hypersensitivity and auto immunity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BERYLLIUM, HLA class II histocompatibility antigen, ... | | Authors: | Clayton, G.M, Crawford, F, Kappler, J.W. | | Deposit date: | 2014-03-12 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of chronic beryllium disease: linking allergic hypersensitivity and autoimmunity.
Cell, 158, 2014
|
|
5LGH
 
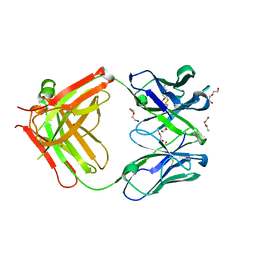 | | Afamin antibody fragment, N14 Fab, L1- glycosilated, crystal form II, same as 5L7X, but isomorphous setting indexed same as 5L88, 5L9D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, MOUSE ANTIBODY FAB FRAGMENT, ... | | Authors: | Rupp, B, Naschberger, A. | | Deposit date: | 2016-07-07 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The N14 anti-afamin antibody Fab: a rare VL1 CDR glycosylation, crystallographic re-sequencing, molecular plasticity and conservative versus enthusiastic modelling.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
8G4G
 
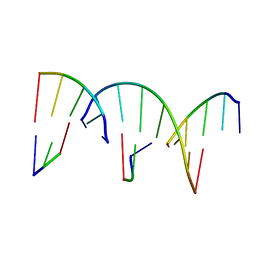 | | Crystal Engineering with One 8-mer DNA | | Descriptor: | DNA (5'-D(*AP*TP*CP*GP*G)-3'), DNA (5'-D(*AP*TP*CP*GP*GP*CP*CP*G)-3'), DNA (5'-D(P*CP*CP*G)-3') | | Authors: | Zhao, J, Zhang, C, Lu, B, Seeman, N.C, Noinaj, N, Sha, R, Mao, C. | | Deposit date: | 2023-02-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Divergence and Convergence: Complexity Emerges in Crystal Engineering from an 8-mer DNA.
J.Am.Chem.Soc., 145, 2023
|
|
4P5M
 
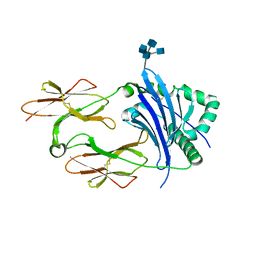 | | Structural Basis of Chronic Beryllium Disease: Bridging the Gap Between Allergic Hypersensitivity and Autoimmunity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)][2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Y, Dai, S, Kappler, J. | | Deposit date: | 2014-03-18 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of chronic beryllium disease: linking allergic hypersensitivity and autoimmunity.
Cell, 158, 2014
|
|
3H0F
 
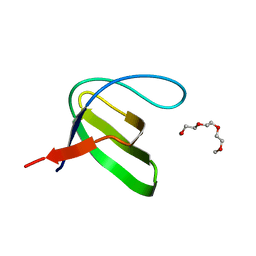 | | Crystal structure of the human Fyn SH3 R96W mutant | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Proto-oncogene tyrosine-protein kinase Fyn | | Authors: | Ponchon, L, Hoh, F, Labesse, G, Dumas, C, Arold, S.T. | | Deposit date: | 2009-04-09 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Synergy and allostery in ligand binding by HIV-1 Nef.
Biochem.J., 478, 2021
|
|
3H0I
 
 | |
1EJ4
 
 | | COCRYSTAL STRUCTURE OF EIF4E/4E-BP1 PEPTIDE | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, EUKARYOTIC INITIATION FACTOR 4E, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E BINDING PROTEIN 1 | | Authors: | Marcotrigiano, J, Gingras, A.-C, Sonenberg, N, Burley, S.K. | | Deposit date: | 2000-02-28 | | Release date: | 2000-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G.
Mol.Cell, 3, 1999
|
|
1GM0
 
 | | A Form of the Pheromone-Binding Protein from Bombyx mori | | Descriptor: | PHEROMONE-BINDING PROTEIN | | Authors: | Horst, R, Damberger, F, Guntert, P, Luginbuhl, P, Nikonova, L, Peng, G, Leal, W.S, Wuthrich, K. | | Deposit date: | 2001-09-05 | | Release date: | 2001-11-30 | | Last modified: | 2018-01-17 | | Method: | SOLUTION NMR | | Cite: | NMR Structure Reveals Novel Intramolecular Regulation Mechanism for Pheromone-Binding and Release
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
6B1E
 
 | | The structure of DPP4 in complex with Vildagliptin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(1r,3s,5R,7S)-3-hydroxytricyclo[3.3.1.1~3,7~]decan-1-yl]amino}-1-{(2S)-2-[(E)-iminomethyl]pyrrolidin-1-yl}ethan-1-o ne, ... | | Authors: | Scapin, G. | | Deposit date: | 2017-09-18 | | Release date: | 2017-09-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A comparative study of the binding properties, dipeptidyl peptidase-4 (DPP-4) inhibitory activity and glucose-lowering efficacy of the DPP-4 inhibitors alogliptin, linagliptin, saxagliptin, sitagliptin and vildagliptin in mice.
Endocrinol Diabetes Metab, 1, 2018
|
|
7Y9N
 
 | | an engineered 5-helix bundle derived from SARS-CoV-2 S2 in complex with HR2P | | Descriptor: | SARS-coV-2 S2 subunit, Spike protein S2',5HB-H2 | | Authors: | Lu, G.W, Lin, X, Guo, L.Y, Lin, S. | | Deposit date: | 2022-06-25 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.885 Å) | | Cite: | An engineered 5-helix bundle derived from SARS-CoV-2 S2 pre-binds sarbecoviral spike at both serological- and endosomal-pH to inhibit virus entry.
Emerg Microbes Infect, 11, 2022
|
|
1DK6
 
 | | NMR structure analysis of the DNA nine base pair duplex D(CATGAGTAC) D(GTAC(NP3)CATG) | | Descriptor: | 5'-D(CP*AP*TP*GP*AP*GP*TP*AP*CP*)-3', 5'-D(GP*TP*AP*CP*(NP3)P*CP*AP*TP*GP*)-3' | | Authors: | Klewer, D.A, Hoskins, A, Davisson, V.J, Bergstrom, D.E, LiWang, A.C. | | Deposit date: | 1999-12-06 | | Release date: | 2000-01-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a DNA duplex containing nucleoside analog 1-(2'-deoxy-beta-D-ribofuranosyl)-3-nitropyrrole and the structure of the unmodified control.
Nucleic Acids Res., 28, 2000
|
|
6B1O
 
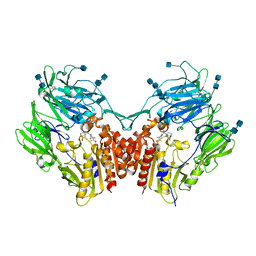 | | The structure of DPP4 in complex with Vildagliptin Analog | | Descriptor: | (2S)-2-amino-1-[(1S,3S,5S)-3-(aminomethyl)-2-azabicyclo[3.1.0]hexan-2-yl]-2-[(1r,3R,5S,7S)-3,5-dihydroxytricyclo[3.3.1.1~3,7~]decan-1-yl]ethan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2017-09-18 | | Release date: | 2017-09-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A comparative study of the binding properties, dipeptidyl peptidase-4 (DPP-4) inhibitory activity and glucose-lowering efficacy of the DPP-4 inhibitors alogliptin, linagliptin, saxagliptin, sitagliptin and vildagliptin in mice.
Endocrinol Diabetes Metab, 1, 2018
|
|
1GL0
 
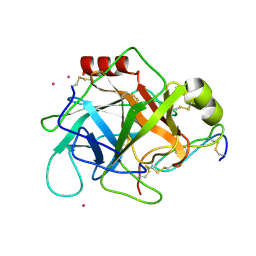 | |
1GL1
 
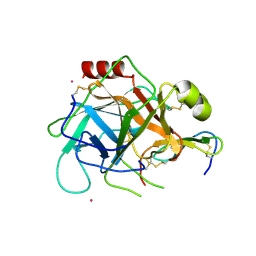 | |
1GQH
 
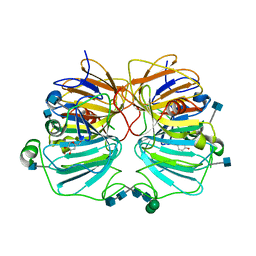 | | Quercetin 2,3-dioxygenase in complex with the inhibitor kojic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-HYDROXY-2-(HYDROXYMETHYL)-4H-PYRAN-4-ONE, ... | | Authors: | Steiner, R.A, Dijkstra, B.W. | | Deposit date: | 2001-11-23 | | Release date: | 2002-06-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional Analysis of the Copper-Dependent Quercetin 2,3-Dioxygenase.1.Ligand-Induced Coordination Changes Probed by X-Ray Crystallography: Inhibition, Ordering Effect and Mechanistic Insights
Biochemistry, 41, 2002
|
|
1GYL
 
 | |
1H1M
 
 | | CRYSTAL STRUCTURE OF QUERCETIN 2,3-DIOXYGENASE ANAEROBICALLY COMPLEXED WITH THE SUBSTRATE KAEMPFEROL | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Steiner, R.A, Dijkstra, B.W. | | Deposit date: | 2002-07-19 | | Release date: | 2002-11-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Anaerobic Enzyme.Substrate Structures Provide Insight Into the Reaction Mechanism of the Copper- Dependent Quercetin 2,3-Dioxygenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1GQG
 
 | | Quercetin 2,3-dioxygenase in complex with the inhibitor diethyldithiocarbamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Steiner, R.A, Dijkstra, B.W. | | Deposit date: | 2001-11-23 | | Release date: | 2002-06-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional Analysis of the Copper-Dependent Quercetin 2,3-Dioxygenase.1.Ligand-Induced Coordination Changes Probed by X-Ray Crystallography: Inhibition, Ordering Effect and Mechanistic Insights
Biochemistry, 41, 2002
|
|
7YKJ
 
 | | Omicron RBDs bound with P3E6 Fab (one up and one down) | | Descriptor: | P3E6 heavy chain, P3E6 light chain, Spike glycoprotein | | Authors: | Tang, B, Dang, S. | | Deposit date: | 2022-07-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into broadly neutralizing antibodies elicited by hybrid immunity against SARS-CoV-2.
Emerg Microbes Infect, 12, 2023
|
|
4M6J
 
 | | Crystal structure of human dihydrofolate reductase (DHFR) bound to NADPH | | Descriptor: | Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.201 Å) | | Cite: | Divergent evolution of protein conformational dynamics in dihydrofolate reductase.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4M6K
 
 | | Crystal structure of human dihydrofolate reductase (DHFR) bound to NADP+ and folate | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, GLYCEROL, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | Divergent evolution of protein conformational dynamics in dihydrofolate reductase.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2HLF
 
 | | Structure of the Escherichis coli ClC chloride channel Y445E mutant and Fab complex | | Descriptor: | BROMIDE ION, Fab Fragment, Heavy chain, ... | | Authors: | Accardi, A, Lobet, S, Williams, C, Miller, C, Dutzler, R. | | Deposit date: | 2006-07-07 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Synergism Between Halide Binding and Proton Transport in a CLC-type Exchanger
J.Mol.Biol., 362, 2006
|
|
