5O75
 
 | | Ube4B U-box domain | | Descriptor: | SULFATE ION, Ubiquitin conjugation factor E4 B | | Authors: | Gabrielsen, M, Buetow, L, Nakasone, M.A, Huang, D.T. | | Deposit date: | 2017-06-08 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.483 Å) | | Cite: | A General Strategy for Discovery of Inhibitors and Activators of RING and U-box E3 Ligases with Ubiquitin Variants.
Mol. Cell, 68, 2017
|
|
5O79
 
 | | Klebsiella pneumoniae OmpK36 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MAGNESIUM ION, OmpK36 | | Authors: | van den berg, B, Pathania, M. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Getting Drugs into Gram-Negative Bacteria: Rational Rules for Permeation through General Porins.
Acs Infect Dis., 4, 2018
|
|
6TDH
 
 | | Crystal structure of Aspergillus fumigatus Glucosamine-6-phosphate N-acetyltransferase 1 in complex with compound 1 | | Descriptor: | 3-(2-chlorophenyl)-4~{H}-1,2,4-triazole, ACETYL COENZYME *A, Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Raimi, O.G, Stanley, M, Lockhart, D. | | Deposit date: | 2019-11-08 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Targeting a critical step in fungal hexosamine biosynthesis.
J.Biol.Chem., 295, 2020
|
|
5OCF
 
 | | Crystal structure of nitric oxide bound to three-domain heme-Cu nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, HEME C, NITRIC OXIDE, ... | | Authors: | Dong, J, Sasaki, D, Eady, R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activation of redox tyrosine switch is required for ligand binding at the catalytic site in heme-cu nitrite reductases
To be published
|
|
6TEP
 
 | | Crystal structure of a galactokinase from Bifidobacterium infantis in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Keenan, T, Parmeggiani, F, Fontenelle, C.Q, Malassis, J, Vendeville, J, Offen, W.A, Both, P, Huang, K, Marchesi, A, Heyam, A, Young, C, Charnock, S, Davies, G.J, Linclau, B, Flitsch, S.L, Fascione, M.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Profiling Substrate Promiscuity of Wild-Type Sugar Kinases for Multi-fluorinated Monosaccharides.
Cell Chem Biol, 27, 2020
|
|
5OCP
 
 | |
5O7U
 
 | |
6TQ3
 
 | |
6TGI
 
 | | Crystal structure of VIM-2 in complex with triazole-based inhibitor OP24 | | Descriptor: | 5-(4-chloranyl-1,5-dimethyl-pyrazol-3-yl)-4-ethyl-1,2,4-triazole-3-thiol, FORMIC ACID, Vim-1, ... | | Authors: | Maso, L, Spirakis, F, Santucci, M, Simon, C, Docquier, J.D, Cruciani, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2019-11-15 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Virtual screening identifies broad-spectrum beta-lactamase inhibitors with activity on clinically relevant serine- and metallo-carbapenemases.
Sci Rep, 10, 2020
|
|
5O9D
 
 | | Crystal structure of R. ruber ADH-A, mutant Y294F, W295A, Y54F, F43H, H39Y | | Descriptor: | (2~{S})-2-methylpentanedioic acid, Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Maurer, D, Hamnevik, E, Enugala, T.R, Widersten, M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Relaxation of nonproductive binding and increased rate of coenzyme release in an alcohol dehydrogenase increases turnover with a nonpreferred alcohol enantiomer.
FEBS J., 284, 2017
|
|
6T3E
 
 | | Structure of Thermococcus litoralis Delta(1)-pyrroline-2-carboxylate reductase in complex with NADH and L-proline | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DELTA1-pyrroline-2-carboxylate reductase, PROLINE | | Authors: | Ferraris, D.M, Miggiano, R, Ferrario, E, Rizzi, M. | | Deposit date: | 2019-10-10 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Thermococcus litoralis Delta1-pyrroline-2-carboxylate reductase in complex with NADH and L-proline.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5OG0
 
 | | Crystal structure of human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) at 2.5 Angstrom; internal aldimine with PLP in the active site | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Borri Voltattorni, C, Cellini, B, Montioli, R. | | Deposit date: | 2017-07-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Radiation damage at the active site of human alanine:glyoxylate aminotransferase reveals that the cofactor position is finely tuned during catalysis.
Sci Rep, 7, 2017
|
|
5OGE
 
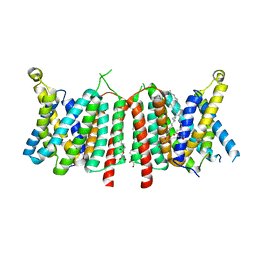 | |
6T40
 
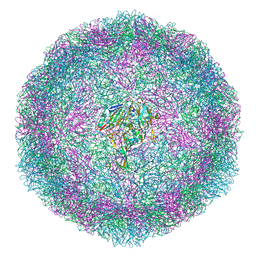 | | Bovine enterovirus F3 in complex with a Cysteinylglycine dipeptide | | Descriptor: | CHLORIDE ION, CYSTEINE, GLYCEROL, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Walter, T.S, Fry, E.E, Stuart, D.I. | | Deposit date: | 2019-10-11 | | Release date: | 2020-08-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Glutathione facilitates enterovirus assembly by binding at a druggable pocket.
Commun Biol, 3, 2020
|
|
6T4V
 
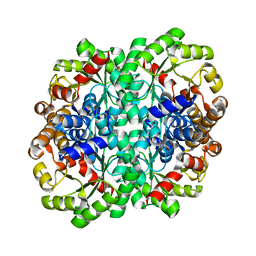 | |
6T5S
 
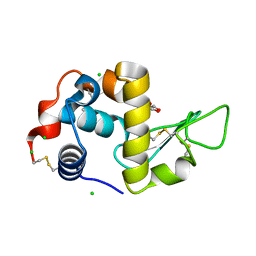 | | Apo form of C-type lysozyme from the upper gastrointestinal tract of Opisthocomus hoatzin | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C | | Authors: | Taylor, E.J, Skjot, M, Skov, L.K, Klausen, M, De Maria, L, Gippert, G.P, Turkenburg, J.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-10-17 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The C-Type Lysozyme from the upper Gastrointestinal Tract of Opisthocomus hoatzin, the Stinkbird.
Int J Mol Sci, 20, 2019
|
|
5OHV
 
 | | K33-specific affimer bound to K33 diUb | | Descriptor: | GLYCEROL, K33-specific affimer, SULFATE ION, ... | | Authors: | Michel, M.A, Komander, D. | | Deposit date: | 2017-07-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Ubiquitin Linkage-Specific Affimers Reveal Insights into K6-Linked Ubiquitin Signaling.
Mol. Cell, 68, 2017
|
|
5OB1
 
 | |
6T6P
 
 | | Crystal structure of Klebsiella pneumoniae FabG2(NADH-dependent) at 1.57 A resolution | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, GLYCEROL, PHOSPHATE ION | | Authors: | Vella, P, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A FabG inhibitor targeting an allosteric binding site inhibits several orthologs from Gram-negative ESKAPE pathogens.
Bioorg.Med.Chem., 30, 2021
|
|
6T7M
 
 | |
6T8E
 
 | |
6T95
 
 | | Trypanothione Reductase from Leismania infantum in complex with 4a | | Descriptor: | 1-[2-[5-[4-(4-azanylbutyl)-3-methyl-1,2,3-triazol-3-ium-1-yl]-2-[4-(2-phenylethyl)-1,3-thiazol-2-yl]phenoxy]ethyl]imidazolidin-2-one, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Carriles, A.A, Hermoso, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of 1,2,3-triazolium salt-based inhibitors of Leishmania infantum trypanothione disulfide reductase with enhanced antileishmanial potency in cellulo and increased selectivity.
Eur.J.Med.Chem., 244, 2022
|
|
5OCM
 
 | | Imine Reductase from Streptosporangium roseum in complex with NADP+ and 2,2,2-trifluoroacetophenone hydrate | | Descriptor: | 2,2,2-trifluoromethyl acetophenone hydrate, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sharma, M, Nestl, B, Grogan, G. | | Deposit date: | 2017-07-03 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | New imine-reducing enzymes from beta-hydroxyacid dehydrogenases by single amino acid substitutions.
Protein Eng. Des. Sel., 31, 2018
|
|
5OKC
 
 | |
5OKN
 
 | | Crystal structure of human SHIP2 Phosphatase-C2 D607A mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Le Coq, J, Lietha, D. | | Deposit date: | 2017-07-25 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for interdomain communication in SHIP2 providing high phosphatase activity.
Elife, 6, 2017
|
|
