1FFS
 
 | | CHEY-BINDING DOMAIN OF CHEA IN COMPLEX WITH CHEY FROM CRYSTALS SOAKED IN ACETYL PHOSPHATE | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, CHEMOTAXIS PROTEIN CHEY, MANGANESE (II) ION | | Authors: | Gouet, P, Chinardet, N, Welch, M, Guillet, V, Birck, C, Mourey, L, Samama, J.-P. | | Deposit date: | 2000-07-26 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Further insights into the mechanism of function of the response regulator CheY from crystallographic studies of the CheY--CheA(124--257) complex.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1FFG
 
 | | CHEY-BINDING DOMAIN OF CHEA IN COMPLEX WITH CHEY AT 2.1 A RESOLUTION | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, CHEMOTAXIS PROTEIN CHEY, MANGANESE (II) ION | | Authors: | Gouet, P, Chinardet, N, Welch, M, Guillet, V, Birck, C, Mourey, L, Samama, J.-P. | | Deposit date: | 2000-07-25 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Further insights into the mechanism of function of the response regulator CheY from crystallographic studies of the CheY--CheA(124--257) complex.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1FFW
 
 | | CHEY-BINDING DOMAIN OF CHEA IN COMPLEX WITH CHEY WITH A BOUND IMIDO DIPHOSPHATE | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, CHEMOTAXIS PROTEIN CHEY, IMIDO DIPHOSPHATE, ... | | Authors: | Gouet, P, Chinardet, N, Welch, M, Guillet, V, Birck, C, Mourey, L, Samama, J.-P. | | Deposit date: | 2000-07-26 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Further insights into the mechanism of function of the response regulator CheY from crystallographic studies of the CheY--CheA(124--257) complex.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
5OAS
 
 | |
1A0O
 
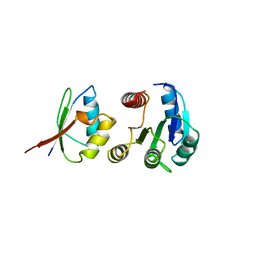 | | CHEY-BINDING DOMAIN OF CHEA IN COMPLEX WITH CHEY | | Descriptor: | CHEA, CHEY, MANGANESE (II) ION | | Authors: | Chinardet, N, Welch, M, Mourey, L, Birck, C, Samama, J.P. | | Deposit date: | 1997-12-05 | | Release date: | 1998-12-30 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the CheY-binding domain of histidine kinase CheA in complex with CheY.
Nat.Struct.Biol., 5, 1998
|
|
5M2E
 
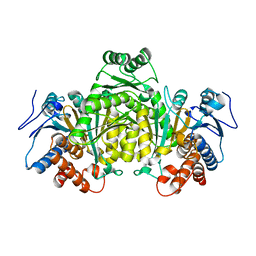 | |
6ZU0
 
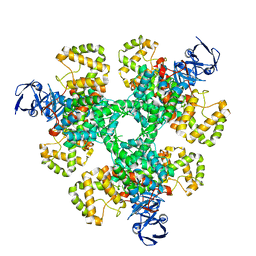 | |
7OO1
 
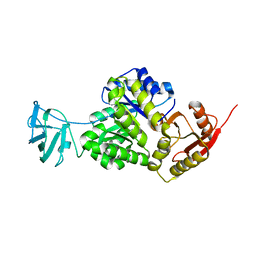 | | Structure, function and characterization of a second pyruvate kinase isozyme in Pseudomonas aeruginosa. | | Descriptor: | Pyruvate kinase | | Authors: | Abdelhamid, Y, Wang, M, Parkhill, S, Brear, P, Welch, M. | | Deposit date: | 2021-05-26 | | Release date: | 2021-11-03 | | Last modified: | 2021-11-24 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structure, Function and Regulation of a Second Pyruvate Kinase Isozyme in Pseudomonas aeruginosa
Front Microbiol, 12, 2021
|
|
6QXL
 
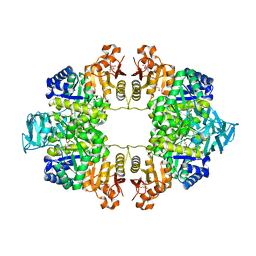 | | Crystal Structure of Pyruvate Kinase II (PykA) from Pseudomonas aeruginosa in complex with sodium malonate, magnesium and glucose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Abdelhamid, Y, Brear, P, Welch, M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Evolutionary plasticity in the allosteric regulator-binding site of pyruvate kinase isoform PykA fromPseudomonas aeruginosa.
J.Biol.Chem., 294, 2019
|
|
6S87
 
 | |
6S62
 
 | |
6S6F
 
 | |
6T5M
 
 | |
6T4V
 
 | |
6G1O
 
 | |
6G3U
 
 | | Structure of Pseudomonas aeruginosa Isocitrate Dehydrogenase, IDH | | Descriptor: | 2-OXOGLUTARIC ACID, Isocitrate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Crousilles, A, Welch, M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.707 Å) | | Cite: | Gluconeogenic precursor availability regulates flux through the glyoxylate shunt inPseudomonas aeruginosa.
J. Biol. Chem., 293, 2018
|
|
