8GMO
 
 | |
8E3N
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with rifamycin S | | Descriptor: | Nuclear receptor subfamily 1 group I member 2, Rifamycin S | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Lin, W, Chen, T. | | Deposit date: | 2022-08-17 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-guided approach to modulate small molecule binding to a promiscuous ligand-activated protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3APM
 
 | | Crystal structure of the human SNP PAD4 protein | | Descriptor: | Protein-arginine deiminase type-4 | | Authors: | Horikoshi, N, Tachiwana, H, Saito, K, Osakabe, A, Sato, M, Yamada, M, Akashi, S, Nishimura, Y, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2010-10-19 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical analyses of the human PAD4 variant encoded by a functional haplotype gene
Acta Crystallogr.,Sect.D, 67, 2011
|
|
7LUK
 
 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C (NHIS-RORGT(244-487)-L6-SRC1(678-692) IN COMPLEX WITH AN AZATRICYCLIC RORGT INVERSE AGONIST | | Descriptor: | (2S)-N-[(6aS,7R,9aS)-9a-[(4-fluorophenyl)sulfonyl]-3-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)-6,6a,7,8,9,9a-hexahydro-5H-cyclopenta[f]quinolin-7-yl]-2-hydroxy-2-methyl-3-(methylsulfonyl)propanamide, Nuclear receptor ROR-gamma | | Authors: | Sack, J.S. | | Deposit date: | 2021-02-22 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.087 Å) | | Cite: | Azatricyclic Inverse Agonists of ROR gamma t That Demonstrate Efficacy in Models of Rheumatoid Arthritis and Psoriasis.
Acs Med.Chem.Lett., 12, 2021
|
|
8DNH
 
 | | Cryo-EM structure of nonmuscle beta-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Arora, A.S, Huang, H.L, Heissler, S.M, Chinthalapudi, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural insights into actin isoforms.
Elife, 12, 2023
|
|
4MD9
 
 | |
1P5F
 
 | | Crystal Structure of Human DJ-1 | | Descriptor: | RNA-binding protein regulatory subunit | | Authors: | Wilson, M.A, Collins, J.L, Hod, Y, Ringe, D, Petsko, G.A. | | Deposit date: | 2003-04-26 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1 A resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early onset Parkinson's disease
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
6H1D
 
 | | Crystal structure of C21orf127-TRMT112 in complex with SAH | | Descriptor: | HemK methyltransferase family member 2, Multifunctional methyltransferase subunit TRM112-like protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wang, S, Hermann, B, Metzger, E, Peng, L, Einsle, O, Schuele, R. | | Deposit date: | 2018-07-11 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | KMT9 monomethylates histone H4 lysine 12 and controls proliferation of prostate cancer cells.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3AQK
 
 | |
3AU5
 
 | |
3SF8
 
 | |
3AQN
 
 | |
3AQL
 
 | |
3BBI
 
 | | Minimally Junctioned Hairpin Ribozyme Incorporating A38(2AP) and A-1 2'-O-Me Modifications near Active Site | | Descriptor: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-11-09 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3BBM
 
 | | Minimally Junctioned Hairpin Ribozyme Incorporates A38C and 2'O-Me Modification at Active Site | | Descriptor: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-11-09 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3BHO
 
 | |
6M95
 
 | | Structure-based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridine-2-one based p38 MAP Kinase Inhibitors by scaffold hopping: compound 1 | | Descriptor: | (4-benzylpiperidin-1-yl)[2-methoxy-4-(methylsulfanyl)phenyl]methanone, Mitogen-activated protein kinase 14 | | Authors: | Lane, W, Okada, K. | | Deposit date: | 2018-08-22 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridin-2-one-Based p38 MAP Kinase Inhibitors: Part 1.
Chemmedchem, 14, 2019
|
|
3B2T
 
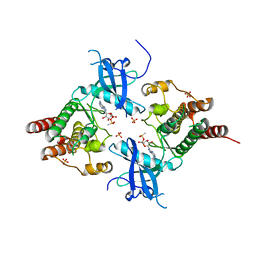 | | Structure of phosphotransferase | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(methyl)phosphoryl]oxy}phosphoryl]adenosine, Fibroblast growth factor receptor 2, PHOSPHATE ION | | Authors: | Lew, E.D, Bae, J.H, Rohmann, E, Wollnik, B, Schlessinger, J. | | Deposit date: | 2007-10-19 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for reduced FGFR2 activity in LADD syndrome: Implications for FGFR autoinhibition and activation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3BAU
 
 | |
1Y9Q
 
 | |
3BAQ
 
 | |
8DK4
 
 | | Peroxisome proliferator-activated receptor gamma in complex with VSP-51-2 | | Descriptor: | 5-(benzylcarbamoyl)-1-[(4-chloro-3-fluorophenyl)methyl]-1H-indole-2-carboxylic acid, Peroxisome proliferator-activated receptor gamma, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | Authors: | Ma, L, Zhou, X.E, Suino-Powell, K, Hou, N, Zhou, Z, Luo, J, Xu, H.E, Yi, W. | | Deposit date: | 2022-07-02 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of VSP-51-2 as the Novel and Safe PPAR gamma Modulator: Structure-Based Design, Biological Validation and Crystal Analysis
To Be Published
|
|
2ZBF
 
 | | Calcium pump crystal structure with bound BeF3 and TG in the absence of calcium | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Toyoshima, C, Ogawa, H, Norimatsu, Y. | | Deposit date: | 2007-10-20 | | Release date: | 2007-12-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | How processing of aspartylphosphate is coupled to lumenal gating of the ion pathway in the calcium pump
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
5ETH
 
 | | RORy in complex with inverse agonist 3. | | Descriptor: | 1-methyl-~{N}-(1-thiophen-2-ylcarbonyl-3,4-dihydro-2~{H}-quinolin-6-yl)-~{N}-[2,2,2-tris(fluoranyl)ethyl]indole-4-sulfonamide, Nuclear receptor ROR-gamma | | Authors: | Marcotte, D.M. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Discovery of biaryls as ROR gamma inverse agonists by using structure-based design.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
2ZH8
 
 | | Complex structure of AFCCA with tRNAminiDGC | | Descriptor: | CCA-adding enzyme, SULFATE ION, tRNA (34-MER) | | Authors: | Toh, Y, Tomita, K. | | Deposit date: | 2008-02-01 | | Release date: | 2008-08-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Molecular basis for maintenance of fidelity during the CCA-adding reaction by a CCA-adding enzyme
Embo J., 27, 2008
|
|
