7NKY
 
 | | RNA Polymerase II-Spt4/5-nucleosome-FACT structure | | Descriptor: | Chromatin elongation factor SPT4, DNA (138-MER), DNA (148-MER), ... | | Authors: | Farnung, L, Ochmann, M, Engeholm, M, Cramer, P. | | Deposit date: | 2021-02-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of nucleosome transcription mediated by Chd1 and FACT.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6MCQ
 
 | | L. pneumophila effector kinase LegK7 in complex with human MOB1A | | Descriptor: | DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, LegK7, ... | | Authors: | Beyrakhova, K.A, Xu, C, Boniecki, M.T, Cygler, M. | | Deposit date: | 2018-09-01 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | TheLegionellakinase LegK7 exploits the Hippo pathway scaffold protein MOB1A for allostery and substrate phosphorylation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7N8N
 
 | | Melbournevirus nucleosome like particle | | Descriptor: | DNA (147-MER), Histone H2B-H2A doublet, Histone H4-H3 doublet | | Authors: | Liu, Y, Toner, C.M, Zhou, K, Bowerman, S, Luger, K. | | Deposit date: | 2021-06-15 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Virus-encoded histone doublets are essential and form nucleosome-like structures.
Cell, 184, 2021
|
|
6M39
 
 | | Cryo-EM structure of SADS-CoV spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ouyang, S, Hongxin, G. | | Deposit date: | 2020-03-03 | | Release date: | 2020-08-26 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Cryo-electron Microscopy Structure of the Swine Acute Diarrhea Syndrome Coronavirus Spike Glycoprotein Provides Insights into Evolution of Unique Coronavirus Spike Proteins.
J.Virol., 94, 2020
|
|
7MZV
 
 | | Structure of yeast pseudouridine synthase 7 (PUS7) | | Descriptor: | Multisubstrate pseudouridine synthase 7, SULFATE ION | | Authors: | Purchal, M, Koutmos, M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Pseudouridine synthase 7 is an opportunistic enzyme that binds and modifies substrates with diverse sequences and structures.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z3T
 
 | | Crystal structure of apo human Cathepsin L | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
8BYQ
 
 | | RNA polymerase II pre-initiation complex with the proximal +1 nucleosome (PIC-Nuc10W) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
7ENA
 
 | | TFIID-based PIC-Mediator holo-complex in pre-assembled state (pre-hPIC-MED) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Qi, Y, Wang, X, Wu, Z, Yin, X, Li, J, Liu, W, Xu, Y. | | Deposit date: | 2021-04-16 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
7ENC
 
 | | TFIID-based PIC-Mediator holo-complex in fully-assembled state (hPIC-MED) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Qi, Y, Wang, X, Wu, Z, Yin, X, Li, J, Liu, W, Xu, Y. | | Deposit date: | 2021-04-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
6CA4
 
 | | Crystal structure of humanized D. rerio TDP2 by 14 mutations | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, MALONATE ION, ... | | Authors: | Shi, K, Aihrara, H. | | Deposit date: | 2018-01-29 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.623 Å) | | Cite: | New fluorescence-based high-throughput screening assay for small molecule inhibitors of tyrosyl-DNA phosphodiesterase 2 (TDP2).
Eur J Pharm Sci, 118, 2018
|
|
7ZS7
 
 | | Crystal structure of human cathepsin L with covalently bound calpain inhibitor VI | | Descriptor: | (2S)-2-[(4-fluorophenyl)sulfonylamino]-3-methyl-N-[(2S)-4-methyl-1-oxidanyl-pentan-2-yl]butanamide, ACETATE ION, Cathepsin L, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 2024
|
|
7TBU
 
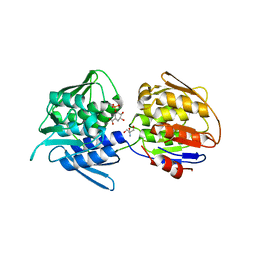 | | Crystal structure of the 5-enolpyruvate-shikimate-3-phosphate synthase (EPSPS) domain of Aro1 from Candida albicans in complex with shikimate-3-phosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-enolpyruvylshikimate-3-phosphate synthase, SHIKIMATE-3-PHOSPHATE | | Authors: | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular analysis and essentiality of Aro1 shikimate biosynthesis multi-enzyme in Candida albicans.
Life Sci Alliance, 5, 2022
|
|
7ZXA
 
 | | Crystal structure of human cathepsin L with covalently bound aloxistatin (E-64D) | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-05-20 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
8CMP
 
 | |
8A4V
 
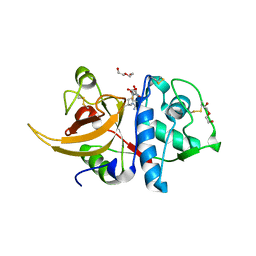 | | Crystal structure of human cathepsin L with covalently bound E-64 | | Descriptor: | Cathepsin L, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
8A4W
 
 | | Crystal structure of human cathepsin L with covalently bound Cathepsin L inhibitor IV | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
8A4X
 
 | | Crystal structure of human Cathepsin L with covalently bound Calpain inhibitor III | | Descriptor: | (phenylmethyl) N-[(2S)-3-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-phenyl-propan-2-yl]amino]butan-2-yl]carbamate, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
8A4U
 
 | | Crystal structure of human cathepsin L with CAA0225 | | Descriptor: | (2S,3S)-N3-[2-(4-hydroxyphenyl)ethyl]-N2-[(2S)-1-oxidanylidene-3-phenyl-1-[(phenylmethyl)amino]propan-2-yl]oxirane-2,3-dicarboxamide, 1,2-ETHANEDIOL, Cathepsin L, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
8A5B
 
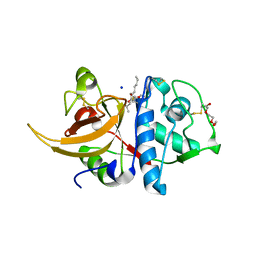 | | Crystal structure of human cathepsin L in complex with covalently bound MG-101 | | Descriptor: | Calpain Inhibitor I, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-06-14 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
7ZVF
 
 | | Crystal structure of human cathepsin L in complex with covalently bound CLIK148 | | Descriptor: | (2S)-N-[(2S)-1-(dimethylamino)-1-oxidanylidene-3-phenyl-propan-2-yl]-2-oxidanyl-N'-(2-pyridin-2-ylethyl)butanediamide, 1,2-ETHANEDIOL, Cathepsin L, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Tsuge, H, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-05-15 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
8JCS
 
 | |
8JCQ
 
 | |
8JCR
 
 | | Crystal structure of Procerain from Calotropis gigantea (pH 6.0) | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, ... | | Authors: | Kumar, A, Jamdar, S.N, Srivastava, G, Makde, R.D. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of Procerain from Calotropis gigantea
To Be Published
|
|
7Z58
 
 | | Crystal structure of human Cathepsin L in complex with covalently bound Calpeptin | | Descriptor: | 1,2-ETHANEDIOL, Calpeptin, Cathepsin L, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
7TOO
 
 | | Yeast 80S ribosome bound with the ALS/FTD-associated dipeptide repeat protein GR20 | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Loveland, A.B, Svidritskiy, E, Susorov, D, Lee, S, Park, A, Zvornicanin, S, Demo, G, Gao, F.B, Korostelev, A.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Ribosome inhibition by C9ORF72-ALS/FTD-associated poly-PR and poly-GR proteins revealed by cryo-EM.
Nat Commun, 13, 2022
|
|
