5APP
 
 | | Actinobacillus actinomycetemcomitans OMP100 residues 133-198 fused to GCN4 adaptors | | Descriptor: | CHLORIDE ION, General control protein GCN4,GENERAL CONTROL PROTEIN GCN4, OUTER MEMBRANE PROTEIN 100,General control protein GCN4 | | Authors: | Hartmann, M.D, Ridderbusch, O, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-09-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | alpha / beta coiled coils.
Elife, 5, 2016
|
|
7QJO
 
 | |
5KHQ
 
 | | Rasip1 RA domain | | Descriptor: | GLYCEROL, Ras-interacting protein 1 | | Authors: | Gingras, A.R. | | Deposit date: | 2016-06-15 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Dimeric Rasip1 RA Domain Recognition of the Ras Subfamily of GTP-Binding Proteins.
Structure, 24, 2016
|
|
5KHO
 
 | | Rasip1 RA domain in complex with Rap1B | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Gingras, A.R. | | Deposit date: | 2016-06-15 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural Basis of Dimeric Rasip1 RA Domain Recognition of the Ras Subfamily of GTP-Binding Proteins.
Structure, 24, 2016
|
|
4O3Z
 
 | | Crystal structure of the vaccine antigen Transferrin Binding Protein B (TbpB) mutant Tyr-95-Ala from Actinobacillus pleuropneumoniae H87 | | Descriptor: | ACETATE ION, GLYCEROL, Outer membrane protein; transferrin-binding protein | | Authors: | Calmettes, C, Yu, R.H, Schryvers, A.B, Moraes, T.F. | | Deposit date: | 2013-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2015-03-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Nonbinding site-directed mutants of transferrin binding protein B exhibit enhanced immunogenicity and protective capabilities.
Infect.Immun., 83, 2015
|
|
4O8M
 
 | | Crystal structure of a trap periplasmic solute binding protein actinobacillus succinogenes 130z, target EFI-510004, with bound L-galactonate | | Descriptor: | CHLORIDE ION, L-galactonic acid, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-28 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4O49
 
 | | Crystal structure of the vaccine antigen Transferrin Binding Protein B (TbpB) mutant Tyr-174-Ala from Actinobacillus pleuropneumoniae H87 | | Descriptor: | ACETATE ION, GLYCEROL, Outer membrane protein; transferrin-binding protein | | Authors: | Calmettes, C, Yu, R.H, Schryvers, A.B, Moraes, T.F. | | Deposit date: | 2013-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2015-03-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nonbinding site-directed mutants of transferrin binding protein B exhibit enhanced immunogenicity and protective capabilities.
Infect.Immun., 83, 2015
|
|
4OGE
 
 | | Crystal structure of the Type II-C Cas9 enzyme from Actinomyces naeslundii | | Descriptor: | HNH endonuclease domain protein, MAGNESIUM ION, SPERMIDINE, ... | | Authors: | Jiang, F, Ma, E, Lin, S, Doudna, J.A. | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structures of Cas9 endonucleases reveal RNA-mediated conformational activation.
Science, 343, 2014
|
|
4OGC
 
 | | Crystal structure of the Type II-C Cas9 enzyme from Actinomyces naeslundii | | Descriptor: | ACETATE ION, HNH endonuclease domain protein, MAGNESIUM ION, ... | | Authors: | Jiang, F, Ma, E, Lin, S, Doudna, J.A. | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-12 | | Last modified: | 2014-03-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Cas9 endonucleases reveal RNA-mediated conformational activation.
Science, 343, 2014
|
|
4O3Y
 
 | | Crystal structure of the vaccine antigen Transferrin Binding Protein B (TbpB) mutant Arg-179-Glu from Actinobacillus pleuropneumoniae H87 | | Descriptor: | ACETATE ION, GLYCEROL, Outer membrane protein; transferrin-binding protein | | Authors: | Calmettes, C, Yu, R.H, Schryvers, A.B, Moraes, T.F. | | Deposit date: | 2013-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2015-03-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nonbinding site-directed mutants of transferrin binding protein B exhibit enhanced immunogenicity and protective capabilities.
Infect.Immun., 83, 2015
|
|
4O3X
 
 | |
1AKP
 
 | | SEQUENTIAL 1H,13C AND 15N NMR ASSIGNMENTS AND SOLUTION CONFORMATION OF APOKEDARCIDIN | | Descriptor: | APOKEDARCIDIN | | Authors: | Constantine, K.L, Colson, K.L, Wittekind, M, Friedrichs, M.S, Zein, N, Tuttle, J, Langley, D.R, Leet, J.E, Schroeder, D.R, Lam, K.S, Farmer II, B.T, Metzler, W.J, Bruccoleri, R.E, Mueller, L. | | Deposit date: | 1994-06-20 | | Release date: | 1994-08-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Sequential 1H, 13C, and 15N NMR assignments and solution conformation of apokedarcidin.
Biochemistry, 33, 1994
|
|
1TXQ
 
 | |
1BHW
 
 | | LOW TEMPERATURE MIDDLE RESOLUTION STRUCTURE OF XYLOSE ISOMERASE FROM MASC DATA | | Descriptor: | XYLOSE ISOMERASE | | Authors: | Ramin, M, Shepard, W, Fourme, R, Kahn, R. | | Deposit date: | 1998-06-10 | | Release date: | 1998-11-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Multiwavelength anomalous solvent contrast (MASC): derivation of envelope structure-factor amplitudes and comparison with model values.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
4A9A
 
 | | Structure of Rbg1 in complex with Tma46 dfrp domain | | Descriptor: | RIBOSOME-INTERACTING GTPASE 1, TRANSLATION MACHINERY-ASSOCIATED PROTEIN 46 | | Authors: | Francis, S.M, Gas, M, Daugeron, M, Seraphin, B, Bravo, J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Rbg1-Tma46 Dimer Structure Reveals New Functional Domains and Their Role in Polysome Recruitment.
Nucleic Acids Res., 40, 2012
|
|
4O3W
 
 | | Crystal structure of the vaccine antigen Transferrin Binding Protein B (TbpB) mutant Tyr-63-Ala from Actinobacillus suis H57 | | Descriptor: | GLYCEROL, SULFATE ION, Transferrin binding protein B | | Authors: | Calmettes, C, Yu, R.H, Schryvers, A.B, Moraes, T.F. | | Deposit date: | 2013-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2015-03-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nonbinding site-directed mutants of transferrin binding protein B exhibit enhanced immunogenicity and protective capabilities.
Infect.Immun., 83, 2015
|
|
8QOY
 
 | | Capsular polysaccharide synthesis multienzyme of Actinobacillus Pleuropneumoniae serotype 3 | | Descriptor: | SULFATE ION, TagF-like capsule polymerase Cps3D, ZINC ION | | Authors: | Di Domenico, V, Litschko, C, Schulze, J, Bethe, A, Cifuente, J.O, Marina, A, Budde, I, Mast, T.A, Sulewska, M, Berger, M, Buettner, F, Gerardy-Schahn, R, Fiebig, T, Guerin, M.E. | | Deposit date: | 2023-09-29 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Transition transferases prime bacterial capsule polymerization
Nat.Chem.Biol., 2024
|
|
5UTT
 
 | | SrtA sortase from Actinomyces oris | | Descriptor: | CHLORIDE ION, Sortase | | Authors: | Osipiuk, J, Ma, X, Ton-That, H, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cell-to-cell interaction requires optimal positioning of a pilus tip adhesin modulated by gram-positive transpeptidase enzymes.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5DDS
 
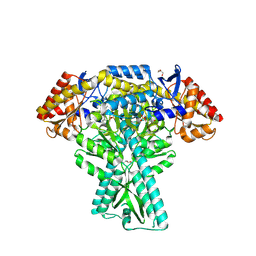 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with PLP | | Descriptor: | ACETIC ACID, CrmG, GLYCEROL, ... | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
5DDW
 
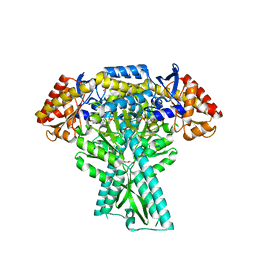 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with the PMP external aldimine adduct with Caerulomycin M | | Descriptor: | CrmG, GLYCEROL, [5-hydroxy-4-({(E)-[(4-hydroxy-2,2'-bipyridin-6-yl)methylidene]amino}methyl)-6-methylpyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
5DDU
 
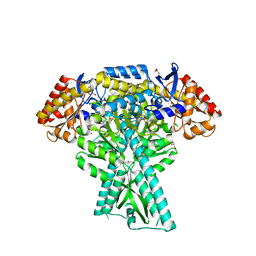 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with PMP | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, CrmG, GLYCEROL, ... | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
3PQS
 
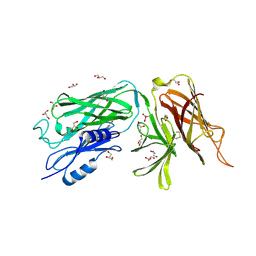 | |
4FZG
 
 | | 20S yeast proteasome in complex with glidobactin | | Descriptor: | Glidobactin, Proteasome component C1, Proteasome component C11, ... | | Authors: | Stein, M, Beck, P, Kaiser, M, Dudler, R, Becker, C.F.W, Groll, M. | | Deposit date: | 2012-07-06 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | One-shot NMR analysis of microbial secretions identifies highly potent proteasome inhibitor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4MFQ
 
 | | The crystal structure of acyltransferase in complex with CoA and 10C-Teicoplanin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose, COENZYME A, ... | | Authors: | Lyu, S.Y, Liu, Y.C, Chang, C.Y, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multiple complexes of long aliphatic N-acyltransferases lead to synthesis of 2,6-diacylated/2-acyl-substituted glycopeptide antibiotics, effectively killing vancomycin-resistant enterococcus
J.Am.Chem.Soc., 136, 2014
|
|
1XG2
 
 | | Crystal structure of the complex between pectin methylesterase and its inhibitor protein | | Descriptor: | Pectinesterase 1, Pectinesterase inhibitor | | Authors: | Di Matteo, A, Raiola, A, Camardella, L, Giovane, A, Bonivento, D, De Lorenzo, G, Cervone, F, Bellincampi, D, Tsernoglou, D. | | Deposit date: | 2004-09-16 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Interaction between Pectin Methylesterase and a Specific Inhibitor Protein
Plant Cell, 17, 2005
|
|
