3J2C
 
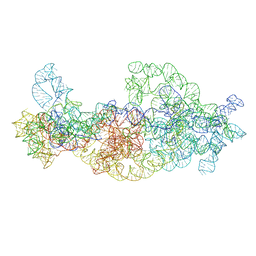 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA body domain, 16S rRNA head domain | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (13.2 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
3J2X
 
 | |
7OL1
 
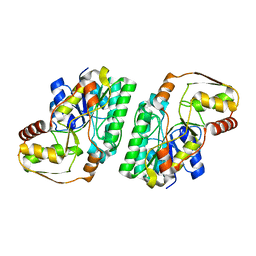 | |
3J3O
 
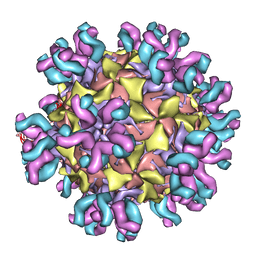 | | Conformational Shift of a Major Poliovirus Antigen Confirmed by Immuno-Cryogenic Electron Microscopy: 160S Poliovirus and C3-Fab Complex | | Descriptor: | C3 antibody, heavy chain, light chain, ... | | Authors: | Lin, J, Cheng, N, Hogle, J.M, Steven, A.C, Belnap, D.M. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.1 Å) | | Cite: | Conformational shift of a major poliovirus antigen confirmed by immuno-cryogenic electron microscopy.
J.Immunol., 191, 2013
|
|
3JD1
 
 | | Glutamate dehydrogenase in complex with NADH, closed conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3VEC
 
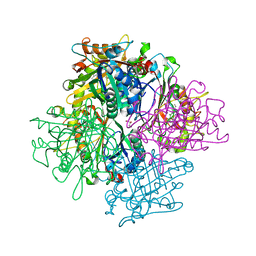 | | Rhodococcus jostii RHA1 DypB D153A variant in complex with heme | | Descriptor: | CHLORIDE ION, DypB, GLYCEROL, ... | | Authors: | Grigg, J.C, Singh, R, Armstrong, Z, Eltis, L.D, Murphy, M.E.P. | | Deposit date: | 2012-01-07 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Distal heme pocket residues of B-type dye-decolorizing peroxidase: arginine but not aspartate is essential for peroxidase activity.
J.Biol.Chem., 287, 2012
|
|
3JD6
 
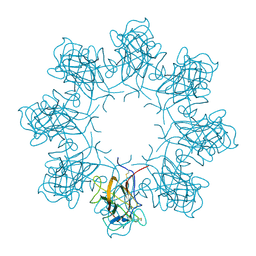 | | Double octamer structure of retinoschisin, a cell-cell adhesion protein of the retina | | Descriptor: | Retinoschisin | | Authors: | Tolun, G, Vijayasarathy, C, Huang, R, Zeng, Y, Li, Y, Steven, A.C, Sieving, P.A, Heymann, J.B. | | Deposit date: | 2016-04-12 | | Release date: | 2016-05-11 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Paired octamer rings of retinoschisin suggest a junctional model for cell-cell adhesion in the retina.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3V9J
 
 | |
3VAI
 
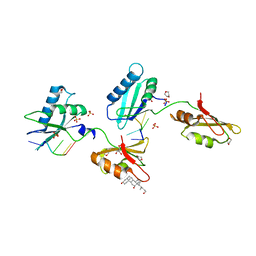 | | Structure of U2AF65 variant with BrU3C5 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA 5'-D(*UP*UP*(BRU)P*UP*CP*UP*U)-3', GLYCEROL, ... | | Authors: | Jenkins, J.L, Frato, K.H, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
3VAS
 
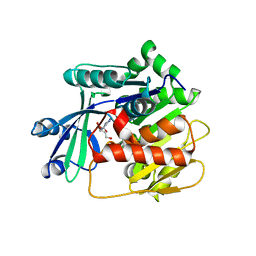 | | Adenosine kinase from Schistosoma mansoni in complex with adenosine in occluded loop conformation | | Descriptor: | ADENOSINE, CHLORIDE ION, Putative adenosine kinase | | Authors: | Romanello, L, Bachega, F.R, Garatt, R.C, DeMarco, R, Brandao-neto, J, Pereira, H.M. | | Deposit date: | 2011-12-29 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Adenosine kinase from Schistosoma mansoni: structural basis for the differential incorporation of nucleoside analogues.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3J3U
 
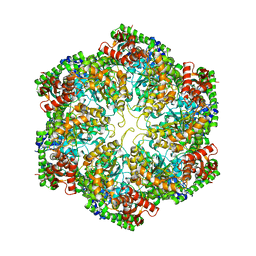 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine.
J.Biol.Chem., 288, 2013
|
|
7OEZ
 
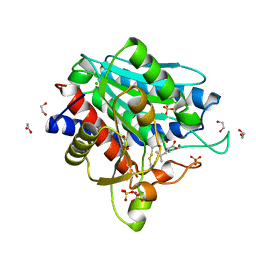 | | Leucine Aminopeptidase A mature enzyme in a complex with leucine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Watson, K.A, Baltulionis, G. | | Deposit date: | 2021-05-04 | | Release date: | 2021-06-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The role of propeptide-mediated autoinhibition and intermolecular chaperone in the maturation of cognate catalytic domain in leucine aminopeptidase.
J.Struct.Biol., 213, 2021
|
|
3VGI
 
 | | The crystal structure of hyperthermophilic family 12 endocellulase from Pyrococcus furiosus | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, Endoglucanase A, ... | | Authors: | Kataoka, M, Kim, H.-W, Ishikawa, K. | | Deposit date: | 2011-08-11 | | Release date: | 2012-09-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Atomic resolution of the crystal structure of the hyperthermophilic family 12 endocellulase and stabilizing role of the DxDxDG calcium-binding motif in Pyrococcus furiosus.
Febs Lett., 586, 2012
|
|
1W49
 
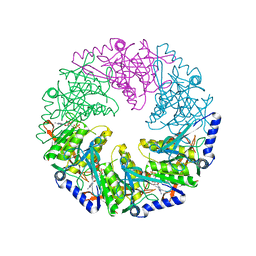 | | P4 protein from Bacteriophage PHI12 in complex with AMPcPP and Mg | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-10-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
3VHA
 
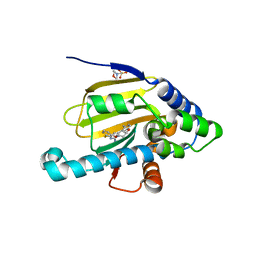 | | Hsp90 alpha N-terminal domain in complex with a macrocyclic inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 22-methyl-13,18-dioxa-7-thia-3,5-diazatetracyclo[17.3.1.1~2,6~.1~8,12~]pentacosa-1(23),2(25),3,5,8(24),9,11,19,21-nonaen-4-amine, Heat shock protein HSP 90-alpha | | Authors: | Fukami, T.A, Ono, N. | | Deposit date: | 2011-08-24 | | Release date: | 2012-07-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Design and synthesis of novel macrocyclic 2-amino-6-arylpyrimidine Hsp90 inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3JQH
 
 | | Structure of the neck region of the glycan-binding receptor DC-SIGNR | | Descriptor: | C-type lectin domain family 4 member M | | Authors: | Feinberg, H, Tso, C.K.W, Taylor, M.E, Drickamer, K, Weis, W.I. | | Deposit date: | 2009-09-06 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Segmented helical structure of the neck region of the glycan-binding receptor DC-SIGNR.
J.Mol.Biol., 394, 2009
|
|
3J68
 
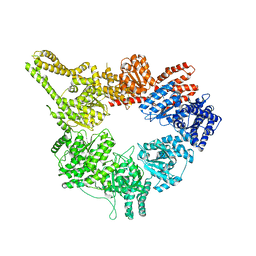 | | Structural mechanism of the dynein powerstroke (pre-powerstroke state) | | Descriptor: | Dynein motor domain | | Authors: | Lin, J, Okada, K, Raytchev, M, Smith, M.C, Nicastro, D. | | Deposit date: | 2013-12-23 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (30 Å) | | Cite: | Structural mechanism of the dynein power stroke.
Nat.Cell Biol., 16, 2014
|
|
3J6T
 
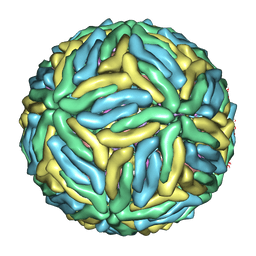 | | Cryo-EM structure of Dengue virus serotype 3 at 37 degrees C | | Descriptor: | envelope protein, membrane protein | | Authors: | Fibriansah, G, Tan, J.L, Smith, S.A, de Alwis, R, Ng, T.-S, Kostyuchenko, V.A, Kukkaro, P, de Silva, A.M, Crowe Jr, J.E, Lok, S.-M. | | Deposit date: | 2014-03-25 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | A highly potent human antibody neutralizes dengue virus serotype 3 by binding across three surface proteins.
Nat Commun, 6, 2015
|
|
3JQK
 
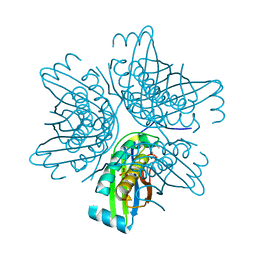 | | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8 (H32 FORM) | | Descriptor: | ACETATE ION, Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION | | Authors: | Kanaujia, S.P, Jeyakanthan, J, Nakagawa, N, Sekar, K, Baba, S, Chen, L, Liu, Z.-J, Wang, B.-C, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-07 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of apo and GTP-bound molybdenum cofactor biosynthesis protein MoaC from Thermus thermophilus HB8
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3JQO
 
 | | Crystal structure of the outer membrane complex of a type IV secretion system | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, LAURYL DIMETHYLAMINE-N-OXIDE, TraF protein, ... | | Authors: | Chandran, V, Fronzes, R, Duquerroy, S, Cronin, N, Navaza, J, Waksman, G. | | Deposit date: | 2009-09-07 | | Release date: | 2009-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the outer membrane complex of a type IV secretion system
Nature, 462, 2009
|
|
3VHK
 
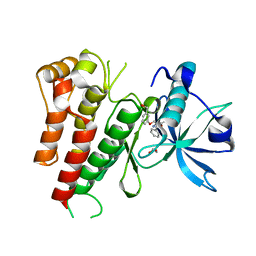 | | Crystal structure of the VEGFR2 kinase domain in complex with a back pocket binder | | Descriptor: | 1,2-ETHANEDIOL, Vascular endothelial growth factor receptor 2, {3-[(5-methyl-2-phenyl-1,3-oxazol-4-yl)methoxy]phenyl}methanol | | Authors: | Iwata, H, Oki, H, Okada, K, Takagi, T, Tawada, M, Miyazaki, Y, Imamura, S, Hori, A, Hixon, M.S, Kimura, H, Miki, H. | | Deposit date: | 2011-08-25 | | Release date: | 2012-09-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | A Back-to-Front Fragment-Based Drug Design Search Strategy Targeting the DFG-Out Pocket of Protein Tyrosine Kinases.
ACS MED.CHEM.LETT., 3, 2012
|
|
3JS6
 
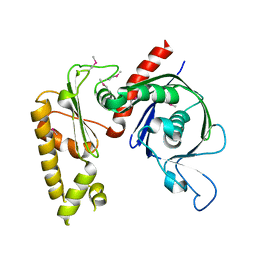 | | Crystal structure of apo psk41 parM protein | | Descriptor: | Uncharacterized ParM protein | | Authors: | Schumacher, M.A, Xu, W, Firth, N. | | Deposit date: | 2009-09-09 | | Release date: | 2010-01-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and filament dynamics of the pSK41 actin-like ParM protein: implications for plasmid DNA segregation.
J.Biol.Chem., 285, 2010
|
|
3VHX
 
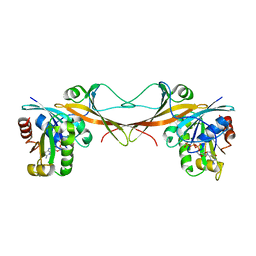 | | The crystal structure of Arf6-MKLP1 (Mitotic kinesin-like protein 1) complex | | Descriptor: | ADP-ribosylation factor 6, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Makyio, H, Takei, T, Ohgi, H, Takahashi, S, Takatsu, H, Ueda, T, Kanaho, Y, Xie, Y, Shin, H.W, Kamikubo, H, Kataoka, M, Kawasaki, M, Kato, R, Wakatsuki, S, Nakayama, K. | | Deposit date: | 2011-09-12 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for Arf6-MKLP1 complex formation on the Flemming body responsible for cytokinesis
Embo J., 31, 2012
|
|
3VCO
 
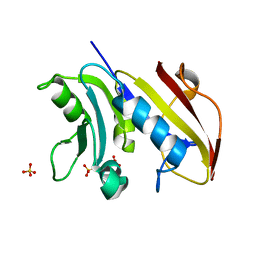 | | Schistosoma mansoni Dihydrofolate reductase | | Descriptor: | Dihydrofolate reductase, SULFATE ION | | Authors: | Serrao, V.H.B, Romanello, L, Cassago, A, DeMarco, R, Pereira, H.M. | | Deposit date: | 2012-01-04 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.946 Å) | | Cite: | Structure and kinetics assays of recombinant Schistosoma mansoni dihydrofolate reductase.
Acta Trop., 170, 2017
|
|
3VDD
 
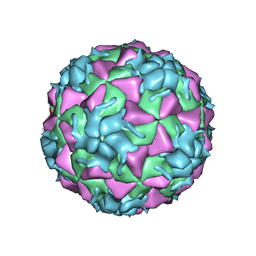 | | Structure of HRV2 capsid complexed with antiviral compound BTA798 | | Descriptor: | 3-ethoxy-6-{2-[1-(6-methylpyridazin-3-yl)piperidin-4-yl]ethoxy}-1,2-benzoxazole, Protein VP1, Protein VP2, ... | | Authors: | Morton, C.J, Feil, S.C, Parker, M.W. | | Deposit date: | 2012-01-05 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | An Orally Available 3-Ethoxybenzisoxazole Capsid Binder with Clinical Activity against Human Rhinovirus.
ACS Med Chem Lett, 3, 2012
|
|
