5KRT
 
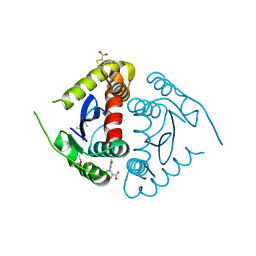 | |
5T3A
 
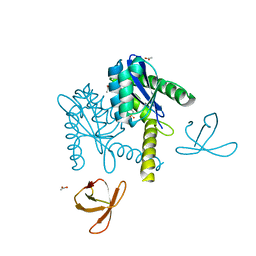 | |
5M0R
 
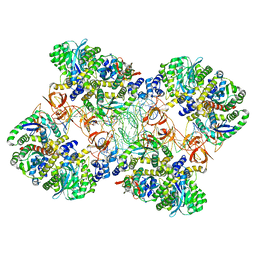 | | Cryo-EM reconstruction of the maedi-visna virus (MVV) strand transfer complex | | Descriptor: | integrase, tDNA, vDNA, ... | | Authors: | Pye, V.E, Ballandras-Colas, A, Maskell, D, Locke, J, Kotecha, A, Costa, A, Cherepanov, P. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | A supramolecular assembly mediates lentiviral DNA integration.
Science, 355, 2017
|
|
5U1C
 
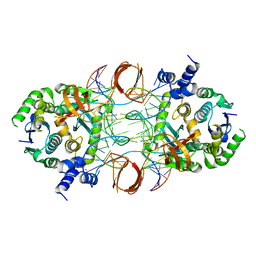 | |
5MMB
 
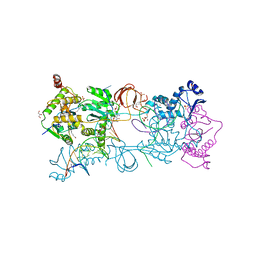 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and the INSTI XZ434 (compound 6p) | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), GLYCEROL, ... | | Authors: | Maskell, D.P, Pye, V.E, Cherepanov, P. | | Deposit date: | 2016-12-09 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure-Guided Optimization of HIV Integrase Strand Transfer Inhibitors.
J. Med. Chem., 60, 2017
|
|
5MMA
 
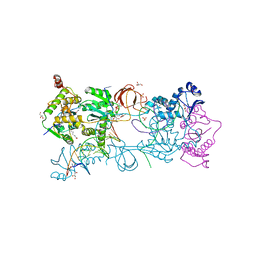 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and the INSTI XZ379 (compound 5'g) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), ... | | Authors: | Maskell, D.P, Pye, V.E, Cherepanov, P. | | Deposit date: | 2016-12-09 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure-Guided Optimization of HIV Integrase Strand Transfer Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UOP
 
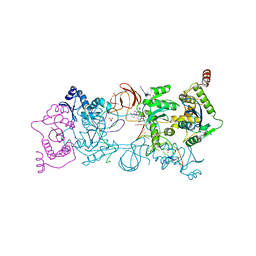 | | CRYSTAL STRUCTURE OF THE PROTOTYPE FOAMY VIRUS INTASOME WITH A 2- PYRIDINONE AMINAL INHIBITOR (COMPOUND 18) | | Descriptor: | (1S,2S,5R)-8'-[(3-chloro-4-fluorophenyl)methyl]-2'-[2-(2,5-dioxo-2,5-dihydro-1H-pyrrol-1-yl)ethyl]-6'-hydroxy-9',10'-dihydro-2'H-spiro[bicyclo[3.1.0]hexane-2,3'-imidazo[5,1-a][2,6]naphthyridine]-1',5',7'(8'H)-trione, GLYCEROL, INTEGRASE, ... | | Authors: | Klein, D.J. | | Deposit date: | 2017-02-01 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery and optimization of 2-pyridinone aminal integrase strand transfer inhibitors for the treatment of HIV.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5UOQ
 
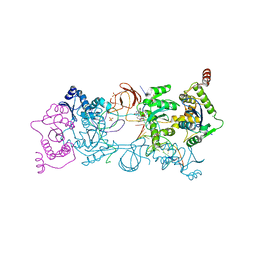 | | CRYSTAL STRUCTURE OF THE PROTOTYPE FOAMY VIRUS INTASOME WITH A 2- PYRIDINONE AMINAL INHIBITOR (COMPOUND 31) | | Descriptor: | (3R)-8-[(3-chloro-4-fluorophenyl)methyl]-6-hydroxy-1,5,7-trioxo-1,2',3',5,7,8,9,10-octahydro-2H-spiro[imidazo[5,1-a][2,6]naphthyridine-3,1'-indene]-7'-carbonitrile, GLYCEROL, INTEGRASE, ... | | Authors: | Klein, D.J. | | Deposit date: | 2017-02-01 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery and optimization of 2-pyridinone aminal integrase strand transfer inhibitors for the treatment of HIV.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5NO1
 
 | |
5OI2
 
 | |
5OI5
 
 | |
5OI3
 
 | |
5OIA
 
 | |
5OI8
 
 | |
6EX9
 
 | |
6EB1
 
 | | HIV-1 Integrase Catalytic Core Domain Complexed with Allosteric Inhibitor (2S)-tert-butoxy[3-(3,4-dihydro-2H-1-benzopyran-6-yl)-1-phenylisoquinolin-4-yl]acetic acid | | Descriptor: | (2S)-tert-butoxy[3-(3,4-dihydro-2H-1-benzopyran-6-yl)-1-phenylisoquinolin-4-yl]acetic acid, Integrase | | Authors: | Lindenberger, J.J, Kobe, M, Kvaratskhelia, M. | | Deposit date: | 2018-08-03 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An Isoquinoline Scaffold as a Novel Class of Allosteric HIV-1 Integrase Inhibitors.
ACS Med Chem Lett, 10, 2019
|
|
6EB2
 
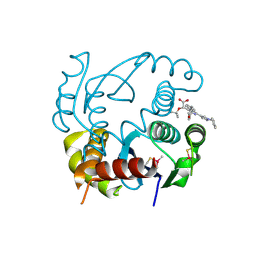 | | HIV-1 Integrase Catalytic Core Domain Complexed with Allosteric Inhibitor (2S)-[1-(1-benzyl-1H-pyrazol-4-yl)-3-(3,4-dihydro-2H-1-benzopyran-6-yl)isoquinolin-4-yl](tert-butoxy)acetic acid | | Descriptor: | (2S)-[1-(1-benzyl-1H-pyrazol-4-yl)-3-(3,4-dihydro-2H-1-benzopyran-6-yl)isoquinolin-4-yl](tert-butoxy)acetic acid, Integrase | | Authors: | Lindenberger, J.J, Kobe, M, Kvaratskhelia, M. | | Deposit date: | 2018-08-03 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | An Isoquinoline Scaffold as a Novel Class of Allosteric HIV-1 Integrase Inhibitors.
ACS Med Chem Lett, 10, 2019
|
|
6NCJ
 
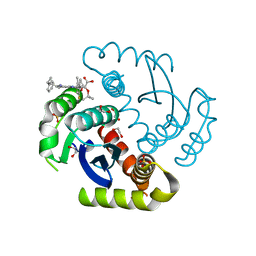 | | Structure of HIV-1 Integrase with potent 5,6,7,8-Tetrahydro-1,6-naphthyridine Derivatives Allosteric Site Inhibitors | | Descriptor: | (2~{S})-2-[4-(8-fluoranyl-5-methyl-3,4-dihydro-2~{H}-chromen-6-yl)-2-methyl-6-[[(1~{S},2~{R})-2-phenylcyclopropyl]methyl]-7,8-dihydro-5~{H}-1,6-naphthyridin-3-yl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, 1,2-ETHANEDIOL, Integrase, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2018-12-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 5,6,7,8-Tetrahydro-1,6-naphthyridine Derivatives as Potent HIV-1-Integrase-Allosteric-Site Inhibitors.
J. Med. Chem., 62, 2019
|
|
6QBT
 
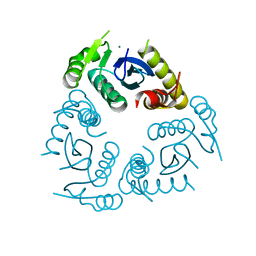 | |
6QBV
 
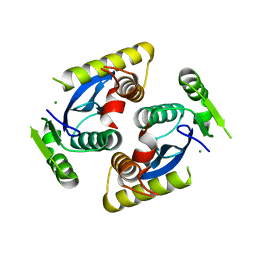 | |
6QBW
 
 | |
6JCG
 
 | | Room temperature structure of HIV-1 Integrase catalytic core domain by serial femtosecond crystallography. | | Descriptor: | CACODYLATE ION, Integrase | | Authors: | Park, J.H, Shi, Y, Han, J, Li, X, Kim, T.H, Yun, J.H. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-Cryogenic Structure and Dynamics of HIV-1 Integrase Catalytic Core Domain by X-ray Free-Electron Lasers.
Int J Mol Sci, 20, 2019
|
|
6JCF
 
 | | Cryogenic structure of HIV-1 Integrase catalytic core domain by synchrotron | | Descriptor: | CACODYLATE ION, Integrase | | Authors: | Park, J.H, Han, J, Kim, T.H, Yun, J.H, Lee, W. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Non-Cryogenic Structure and Dynamics of HIV-1 Integrase Catalytic Core Domain by X-ray Free-Electron Lasers.
Int J Mol Sci, 20, 2019
|
|
6NUJ
 
 | |
6RNY
 
 | | PFV intasome - nucleosome strand transfer complex | | Descriptor: | DNA (108-MER), DNA (128-MER), DNA (33-MER), ... | | Authors: | Pye, V.E, Renault, L, Maskell, D.P, Cherepanov, P, Costa, A. | | Deposit date: | 2019-05-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Retroviral integration into nucleosomes through DNA looping and sliding along the histone octamer.
Nat Commun, 10, 2019
|
|
