8RCT
 
 | |
7QK5
 
 | |
5DYW
 
 | | Crystal structure of human butyrylcholinesterase in complex with N-((1-benzylpiperidin-3-yl)methyl)-N-(2-methoxyethyl)naphthalene-2-sulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Coquelle, N, Brus, B, Colletier, J.P. | | Deposit date: | 2015-09-25 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of an in-vivo active reversible butyrylcholinesterase inhibitor.
Sci Rep, 6, 2016
|
|
8RGP
 
 | | Closed Complex I from murine brain | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2023-12-14 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | SCAF1 drives the compositional diversity of mammalian respirasomes.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7QSD
 
 | | Bovine complex I in the active state at 3.1 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Bridges, H.R, Blaza, J.N, Yin, Z, Chung, I, Hirst, J. | | Deposit date: | 2022-01-13 | | Release date: | 2022-03-02 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of mammalian respiratory complex I inhibition by medicinal biguanides.
Science, 379, 2023
|
|
8EXH
 
 | | Agrobacterium tumefaciens Tpilus | | Descriptor: | (14S,17R)-20-amino-17-hydroxy-11,17-dioxo-12,16,18-trioxa-17lambda~5~-phosphaicosan-14-yl tetradecanoate, Protein virB2 | | Authors: | Beltran, L.C, Egelman, E.H. | | Deposit date: | 2022-10-25 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Archaeal DNA-import apparatus is homologous to bacterial conjugation machinery
Nat Commun, 14, 2023
|
|
8EN5
 
 | | Structure of GII.4 norovirus in complex with Nanobody 56 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GII.4 P domain, ... | | Authors: | Kher, G, Sabin, C, Pancera, M, Koromyslova, A, Hansman, G. | | Deposit date: | 2022-09-28 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Direct Blockade of the Norovirus Histo-Blood Group Antigen Binding Pocket by Nanobodies.
J.Virol., 97, 2023
|
|
8QR0
 
 | | Cryo-EM structure of the light-driven sodium pump ErNaR in the pentameric form at pH 4.3 | | Descriptor: | Bacteriorhodopsin-like protein, DODECYL-BETA-D-MALTOSIDE, EICOSANE | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Marin, E, Stetsenko, A, Guskov, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
7QH6
 
 | | Cryo-EM structure of the human mtLSU assembly intermediate upon MRM2 depletion - class 1 | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L13, mitochondrial, ... | | Authors: | Rebelo-Guiomar, P, Pellegrino, S, Dent, K.C, Warren, A.J, Minczuk, M. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-02 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | A late-stage assembly checkpoint of the human mitochondrial ribosome large subunit.
Nat Commun, 13, 2022
|
|
7QIQ
 
 | | CRYSTAL STRUCTURE OF THE P1 aminobutanoic acid (ABU) BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Wahl, M.C, Loll, B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fluorine-induced polarity increases inhibitory activity of BPTI towards chymotrypsin.
Rsc Chem Biol, 3, 2022
|
|
8F4F
 
 | | RT XFEL structure of Photosystem II 500 microseconds after the third illumination at 2.03 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4H
 
 | | RT XFEL structure of Photosystem II 1200 microseconds after the third illumination at 2.10 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
7QWR
 
 | | Structure of the ribosome-nascent chain containing an ER signal sequence in complex with NAC | | Descriptor: | 28S rRNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Jomaa, A, Gamerdinger, M, Hsieh, H, Wallisch, A, Chandrasekaran, V, Ulusoy, Z, Scaiola, A, Hegde, R, Shan, S, Ban, N, Deuerling, E. | | Deposit date: | 2022-01-25 | | Release date: | 2022-03-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting.
Science, 375, 2022
|
|
8EZ5
 
 | | RT XFEL structure of the two-flash state of Photosystem II (2F, S3-rich) at 2.09 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-10-31 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8FRH
 
 | |
8F4C
 
 | | RT XFEL structure of the two-flash state of Photosystem II (2F, S3-rich) at 2.00 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8QAX
 
 | | Medicago truncatula HISN5 (IGPD) in complex with MN, FMT, GOL and TRS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, GLYCEROL, ... | | Authors: | Witek, W, Ruszkowski, M. | | Deposit date: | 2023-08-23 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Targeting imidazole-glycerol phosphate dehydratase in plants: novel approach for structural and functional studies, and inhibitor blueprinting.
Front Plant Sci, 15, 2024
|
|
8F4I
 
 | | RT XFEL structure of Photosystem II 2000 microseconds after the third illumination at 2.00 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8QO9
 
 | | Cryo-EM structure of a human spliceosomal B complex protomer | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, MINX pre-mRNA, Microfibrillar-associated protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2023-09-28 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (5.29 Å) | | Cite: | Cryo-EM analyses of dimerized spliceosomes provide new insights into the functions of B complex proteins.
Embo J., 43, 2024
|
|
7QWS
 
 | | Structure of ribosome translating beta-tubulin in complex with TTC5 and NAC | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Jomaa, A, Gamerdinger, M, Hsieh, H, Wallisch, A, Chandrasekaran, V, Ulusoy, Z, Scaiola, A, Hegde, R, Shan, S, Ban, N, Deuerling, E. | | Deposit date: | 2022-01-25 | | Release date: | 2022-03-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting.
Science, 375, 2022
|
|
8QYF
 
 | |
7QNQ
 
 | | Structure of the Aux2 relaxosome protein of plasmid pLS20 | | Descriptor: | Auxiliary relaxosome protein, CHLORIDE ION, MAGNESIUM ION | | Authors: | Boer, D.R, Crespo, I. | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and biochemical characterization of the relaxosome auxiliary proteins encoded on the Bacillus subtilis plasmid pLS20.
Comput Struct Biotechnol J, 20, 2022
|
|
8F4K
 
 | | RT XFEL structure of the three-flash state of Photosystem II (3F, S0-rich) at 2.16 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
6O2B
 
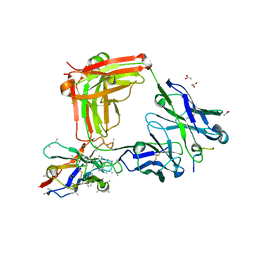 | | Crystal structure of 4493 Fab in complex with circumsporozoite protein DND and anti-kappa VHH domain | | Descriptor: | 1,2-ETHANEDIOL, 4493 Fab heavy chain, 4493 Kappa light chain, ... | | Authors: | Scally, S.W, Bosch, A, Prieto, K, Murugan, R, Wardemann, H, Julien, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of protective human antibodies against Plasmodium falciparum circumsporozoite protein repeat motifs.
Nat. Med., 26, 2020
|
|
7QY3
 
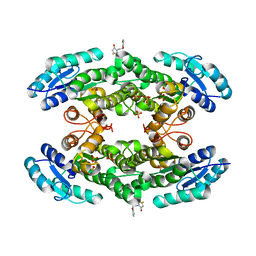 | | Crystal structure of the halohydrin dehalogenase HheG D114C mutant cross-linked with BMOE | | Descriptor: | 1,1'-ethane-1,2-diylbis(1H-pyrrole-2,5-dione), Putative oxidoreductase, SULFATE ION | | Authors: | Henke, S, Blankenfeldt, W, Schallmey, A. | | Deposit date: | 2022-01-27 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Biocatalytically active and stable cross-linked enzyme crystals of halohydrin dehalogenase HheG by protein engineering
Chemcatchem, 2022
|
|
