7MFS
 
 | |
7MSK
 
 | | ThuS glycosin S-glycosyltransferase | | Descriptor: | Glyco_trans_2-like domain-containing protein, MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUORO-ALPHA-D-GLUCOSE | | Authors: | Garg, N, Nair, S.K. | | Deposit date: | 2021-05-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and mechanistic investigations of protein S-glycosyltransferases.
Cell Chem Biol, 28, 2021
|
|
8TY3
 
 | | MI-31 ligand bound to SARS-CoV-2 Mpro | | Descriptor: | (1S,3aR,6aS)-2-[(3,4-dichlorophenoxy)acetyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase nsp5 | | Authors: | Blankenship, L.R, Liu, W.R. | | Deposit date: | 2023-08-24 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | MI-31 bound to SARS-CoV-2 Mpro
To Be Published
|
|
6DXH
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with 4-(4-tert-butylphenyl)-4-oxobutanoate | | Descriptor: | 4-(4-tert-butylphenyl)-4-oxobutanoic acid, UNKNOWN ATOM OR ION, Ubiquitin carboxyl-terminal hydrolase 5, ... | | Authors: | Harding, R.J, Mann, M.K, Ravichandran, M, Ferreira de Freitas, R, Franzoni, I, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Schapira, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-28 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Small Molecule Antagonists of the USP5 Zinc Finger Ubiquitin-Binding Domain.
J.Med.Chem., 62, 2019
|
|
8TY4
 
 | | MI-30 bound to Mpro of SARS-CoV-2 | | Descriptor: | (1S,3aR,6aS)-2-[(2,4-dichlorophenoxy)acetyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase nsp5 | | Authors: | Blankenship, L.R, Liu, W.R. | | Deposit date: | 2023-08-24 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | MI-30 bound to SARS-CoV-2 Mpro
To Be Published
|
|
8TZI
 
 | | Human equilibrative nucleoside transporter-1, JH-ENT-01 bound | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[4-[3-[3,4-dimethoxy-5-[[4-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]methoxy]phenyl]carbonyloxypropyl]-1,4-diazepan-1-yl]propyl 3,4,5-trimethoxybenzoate, Equilibrative nucleoside transporter 1 | | Authors: | Wright, N.J, Lee, S.Y. | | Deposit date: | 2023-08-26 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Rational ENT1 inhibition as a pathway for neuropathic pain relief
To Be Published
|
|
6HLP
 
 | | Crystal structure of the Neurokinin 1 receptor in complex with the small molecule antagonist Netupitant | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-[3,5-bis(trifluoromethyl)phenyl]-~{N},2-dimethyl-~{N}-[4-(2-methylphenyl)-6-(4-methylpiperazin-1-yl)pyridin-3-yl]propanamide, CITRIC ACID, ... | | Authors: | Schoppe, J, Ehrenmann, J, Klenk, C, Rucktooa, P, Schutz, M, Dore, A.S, Pluckthun, A. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the human neurokinin 1 receptor in complex with clinically used antagonists.
Nat Commun, 10, 2019
|
|
6HSY
 
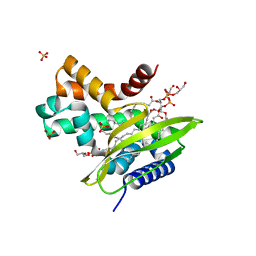 | | Two-phospholipid-bound crystal structure of the substrate-binding protein Ttg2D from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, Phospholipid PG(16:0/cy17:0), SULFATE ION, ... | | Authors: | Costenaro, L, Conchillo-Sole, O, Daura, X. | | Deposit date: | 2018-10-02 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The Pseudomonas aeruginosa substrate-binding protein Ttg2D functions as a general glycerophospholipid transporter across the periplasm.
Commun Biol, 4, 2021
|
|
6DXT
 
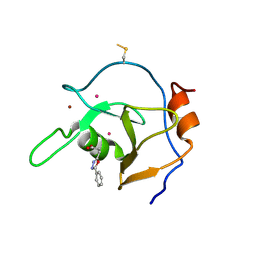 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with 3-(5-phenyl-1,3,4-oxadiazol-2-yl)propanoate | | Descriptor: | 1,2-ETHANEDIOL, 3-(5-phenyl-1,3,4-oxadiazol-2-yl)propanoic acid, UNKNOWN ATOM OR ION, ... | | Authors: | Mann, M.K, Harding, R.J, Ravichandran, M, Ferreira de Freitas, R, Franzoni, I, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Schapira, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-29 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of Small Molecule Antagonists of the USP5 Zinc Finger Ubiquitin-Binding Domain.
J.Med.Chem., 62, 2019
|
|
7MQT
 
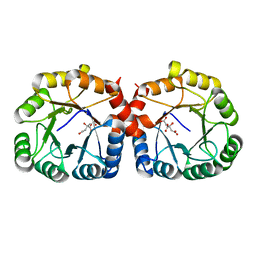 | |
8TWY
 
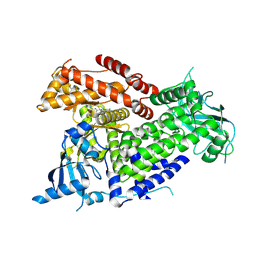 | | Structure of PIK3CA with covalent inhibitor ALO26 | | Descriptor: | 1-(4-{[2-(4-{(4P)-4-[2-amino-4-(difluoromethyl)pyrimidin-5-yl]-6-[(3S)-3-methylmorpholin-4-yl]-1,3,5-triazin-2-yl}piperazin-1-yl)-2-oxoethoxy]methyl}piperidin-1-yl)prop-2-en-1-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Burke, J.E, Barlow-Busch, I. | | Deposit date: | 2023-08-21 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structure of PIK3CA with covalent inhibitor ALO26
To Be Published
|
|
8B61
 
 | | Crystal structure of BfrC protein from Bacteroides fragilis NCTC 9343 | | Descriptor: | Conserved hypothetical lipoprotein, GLYCEROL, pentane-1,3,5-tricarboxylic acid | | Authors: | Antonyuk, S.V, Barnett, K, Strange, R.W, Olczak, T. | | Deposit date: | 2022-09-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Bacteroides fragilis expresses three proteins similar to Porphyromonas gingivalis HmuY: Hemophore-like proteins differentially evolved to participate in heme acquisition in oral and gut microbiomes.
Faseb J., 37, 2023
|
|
7MGQ
 
 | | AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans | | Descriptor: | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase, MAGNESIUM ION | | Authors: | Wizrah, M.S, Chua, S.M.H, Luo, Z, Manik, M.K, Pan, M, Whyte, J.M, Robertson, A.B, Kappler, U, Kobe, B, Fraser, J.A. | | Deposit date: | 2021-04-13 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans.
J.Biol.Chem., 298, 2022
|
|
8B6A
 
 | |
8U05
 
 | |
8U0I
 
 | | Crystal structure of PA0012 complexed with cyclic-di-GMP from Pseudomonas aeruginosa | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), GLYCEROL, PilZ domain-containing protein | | Authors: | Hammons, N.A, Schnicker, N.J, Fuentes, E.J. | | Deposit date: | 2023-08-29 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | Crystal structure of PA0012 complexed with cyclic-di-GMP from Pseudomonas aeruginosa
To Be Published
|
|
7MFN
 
 | |
6DE5
 
 | |
8U07
 
 | |
8BCS
 
 | | X-ray crystal structure of a de novo designed helix-loop-helix homodimer in an anti arrangement, CC-HP1.0 | | Descriptor: | ACETATE ION, CC-HP1.0 | | Authors: | Edgell, C.L, Mylemans, B, Naudin, E.A, Smith, A.J, Savery, N.J, Woolfson, D.N. | | Deposit date: | 2022-10-17 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and Selection of Heterodimerizing Helical Hairpins for Synthetic Biology.
Acs Synth Biol, 12, 2023
|
|
6HMU
 
 | | Ternary complex of Estrogen Receptor alpha peptide and 14-3-3 sigma C45 mutant bound to disulfide fragment PPI stabilizer 6 | | Descriptor: | 14-3-3 protein sigma, 2-(4-chloranylphenoxy)-2-methyl-~{N}-(3-sulfanylpropyl)propanamide, Estrogen receptor, ... | | Authors: | Sijbesma, E, Hallenbeck, K.K, Leysen, S, Arkin, M.R, Ottmann, C. | | Deposit date: | 2018-09-12 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Site-Directed Fragment-Based Screening for the Discovery of Protein-Protein Interaction Stabilizers.
J. Am. Chem. Soc., 141, 2019
|
|
7MHW
 
 | | Crystal structure of the protease inhibitor U-Omp19 from Brucella abortus fused to Maltose-binding protein | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Outer membrane lipoprotein omp19, SULFATE ION | | Authors: | Darriba, M.L, Klinke, S, Otero, L.H, Cerutti, M.L, Cassataro, J, Pasquevich, K.A. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A disordered region retains the full protease inhibitor activity and the capacity to induce CD8 + T cells in vivo of the oral vaccine adjuvant U-Omp19.
Comput Struct Biotechnol J, 20, 2022
|
|
8T7N
 
 | | Crystal structure of the R132H mutant of IDH1 bound to compound 1 | | Descriptor: | Isocitrate dehydrogenase [NADP] cytoplasmic, N-(4-tert-butylphenyl)-7,8-dimethyl-5,11-dihydro-6H-pyrido[2,3-b][1,5]benzodiazepine-6-carboxamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lu, J, Abeywickrema, P, Heo, M.R, Parthasarathy, G, McCoy, M, Soisson, S.M. | | Deposit date: | 2023-06-20 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Mechanistic and Biostructural Studies of Mutant IDH1
To Be Published
|
|
6DJX
 
 | | Crystal Structure of pParkin-pUb-UbcH7 complex | | Descriptor: | RBR-type E3 ubiquitin transferase,RBR-type E3 ubiquitin transferase, Ubiquitin, Ubiquitin-conjugating enzyme E2 L3, ... | | Authors: | Sauve, V, Sung, G, Trempe, J.F, Gehring, K. | | Deposit date: | 2018-05-27 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.801 Å) | | Cite: | Mechanism of parkin activation by phosphorylation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7MHD
 
 | | Thioesterase Domain of Human Fatty Acid Synthase (FASN-TE) binding a competitive inhibitor SBP-7635 | | Descriptor: | Fatty acid synthase, N,N-diethyl-4-{2-[(2-fluorophenyl)methyl]-1,3-thiazol-4-yl}benzene-1-sulfonamide | | Authors: | Aleshin, A.E, Lambert, L, Liddington, R.C, Cosford, N. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Thioesterase Domain of Human Fatty Acid Synthase (FASN-TE) binding a competitive inhibitor SBP-7635
To Be Published
|
|
