3PHM
 
 | |
4IES
 
 | | Cys-persulfenate bound Cysteine Dioxygenase at pH 6.2 in the presence of Cys | | Descriptor: | Cysteine dioxygenase type 1, FE (III) ION, S-HYDROPEROXYCYSTEINE | | Authors: | Driggers, C.M, Cooley, R.B, Sankaran, B, Karplus, P.A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cysteine Dioxygenase Structures from pH4 to 9: Consistent Cys-Persulfenate Formation at Intermediate pH and a Cys-Bound Enzyme at Higher pH.
J.Mol.Biol., 425, 2013
|
|
1R33
 
 | | Golgi alpha-mannosidase II complex with 5-thio-D-mannopyranosylamine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-thio-alpha-D-mannopyranosylamine, ... | | Authors: | Kuntz, D.A, Xin, W, Kavelekar, L.M, Rose, D.R, Pinto, B.M. | | Deposit date: | 2003-09-30 | | Release date: | 2004-11-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 5-Thio-d-glycopyranosylamines and their amidinium salts as potential transition-state mimics of glycosyl hydrolases: synthesis, enzyme inhibitory activities, X-ray crystallography, and molecular modeling
TETRAHEDRON ASYMMETRY, 16, 2005
|
|
6RYN
 
 | |
3NEY
 
 | | Crystal structure of the kinase domain of MPP1/p55 | | Descriptor: | 55 kDa erythrocyte membrane protein, SULFATE ION, UNKNOWN ATOM OR ION | | Authors: | Shen, Y, Tong, Y, Zhong, N, Guan, X, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of the kinase domain of MPP1/p55
To be Published
|
|
1R46
 
 | | Structure of human alpha-galactosidase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Garman, S.C, Garboczi, D.N. | | Deposit date: | 2003-10-03 | | Release date: | 2004-03-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The molecular defect leading to Fabry disease: structure of human alpha-galactosidase
J.Mol.Biol., 337, 2004
|
|
3R4L
 
 | | Human very long half life Plasminogen Activator Inhibitor type-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plasminogen activator inhibitor 1, beta-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yang, J, Zheng, H, Han, Q, Skrzypczak-Jankun, E, Jankun, J. | | Deposit date: | 2011-03-17 | | Release date: | 2011-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Remarkable extension of PAI-1 half-life surprisingly brings no changes to its structure.
Int.J.Mol.Med., 29, 2012
|
|
3R4Z
 
 | | Crystal structure of alpha-neoagarobiose hydrolase (ALPHA-NABH) in complex with alpha-d-galactopyranose from Saccharophagus degradans 2-40 | | Descriptor: | Glycosyl hydrolase family 32, N terminal, alpha-D-galactopyranose | | Authors: | Lee, S, Lee, J.Y, Ha, S.C, Shin, D.H, Kim, K.H, Bang, W.G, Kim, S.H, Choi, I.G. | | Deposit date: | 2011-03-18 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of a key enzyme in the agarolytic pathway, alpha-neoagarobiose hydrolase from Saccharophagus degradans 2-40
Biochem.Biophys.Res.Commun., 412, 2011
|
|
4II8
 
 | | Lysozyme with Benzyl alcohol | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Sharma, P, Ashish | | Deposit date: | 2012-12-20 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Characterization of heat induced spherulites of lysozyme reveals new insight on amyloid initiation
Sci Rep, 6, 2016
|
|
4IIH
 
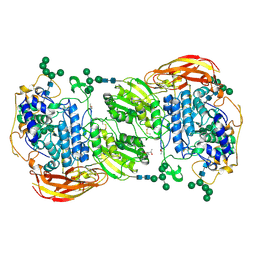 | | Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with thiocellobiose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Suzuki, K, Sumitani, J, Kawaguchi, T, Fushinobu, S. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of glycoside hydrolase family 3 beta-glucosidase 1 from Aspergillus aculeatus
Biochem.J., 452, 2013
|
|
3N81
 
 | |
1BIX
 
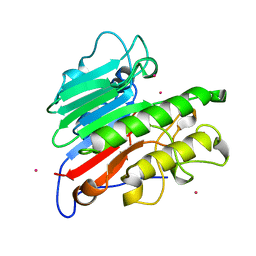 | | THE CRYSTAL STRUCTURE OF THE HUMAN DNA REPAIR ENDONUCLEASE HAP1 SUGGESTS THE RECOGNITION OF EXTRA-HELICAL DEOXYRIBOSE AT DNA ABASIC SITES | | Descriptor: | AP ENDONUCLEASE 1, PLATINUM (II) ION, SAMARIUM (III) ION | | Authors: | Gorman, M.A, Morera, S, Rothwell, D.G, De La Fortelle, E, Mol, C.D, Tainer, J.A, Hickson, I.D, Freemont, P.S. | | Deposit date: | 1998-06-19 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites.
EMBO J., 16, 1997
|
|
4IEM
 
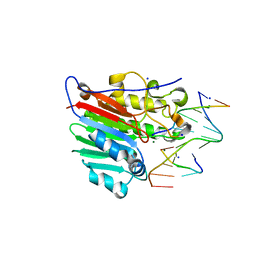 | | Human apurinic/apyrimidinic endonuclease (APE1) with product DNA and Mg2+ | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*GP*GP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*C)-3'), DNA (5'-D(P*(3DR)P*GP*AP*TP*CP*G)-3'), ... | | Authors: | Tsutakawa, S.E, Mol, C.D, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3936 Å) | | Cite: | Conserved Structural Chemistry for Incision Activity in Structurally Non-homologous Apurinic/Apyrimidinic Endonuclease APE1 and Endonuclease IV DNA Repair Enzymes.
J.Biol.Chem., 288, 2013
|
|
3R8W
 
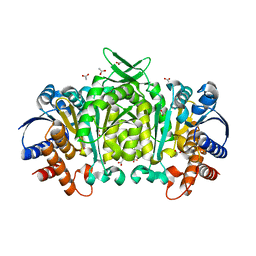 | | Structure of 3-isopropylmalate dehydrogenase isoform 2 from Arabidopsis thaliana at 2.2 angstrom resolution | | Descriptor: | 3-isopropylmalate dehydrogenase 2, chloroplastic, ACETATE ION | | Authors: | He, Y, Galant, A, Pang, Q, Strul, J.M, Balogun, S, Jez, J.M, Chen, S. | | Deposit date: | 2011-03-24 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional evolution of isopropylmalate dehydrogenases in the leucine and glucosinolate pathways of Arabidopsis thaliana.
J.Biol.Chem., 286, 2011
|
|
3R31
 
 | |
3NK4
 
 | |
3N9Y
 
 | | Crystal structure of human CYP11A1 in complex with cholesterol | | Descriptor: | Adrenodoxin, CHOLESTEROL, Cholesterol side-chain cleavage enzyme, ... | | Authors: | Strushkevich, N.V, MacKenzie, F, Tempel, W, Botchkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J.U, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-31 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for pregnenolone biosynthesis by the mitochondrial monooxygenase system.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4IIB
 
 | | Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Suzuki, K, Sumitani, J, Kawaguchi, T, Fushinobu, S. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of glycoside hydrolase family 3 beta-glucosidase 1 from Aspergillus aculeatus
Biochem.J., 452, 2013
|
|
1BVU
 
 | | GLUTAMATE DEHYDROGENASE FROM THERMOCOCCUS LITORALIS | | Descriptor: | PROTEIN (GLUTAMATE DEHYDROGENASE) | | Authors: | Baker, P.J, Britton, K.L, Yip, K.S, Stillman, T.J, Rice, D.W. | | Deposit date: | 1999-07-20 | | Release date: | 1999-09-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure determination of the glutamate dehydrogenase from the hyperthermophile Thermococcus litoralis and its comparison with that from Pyrococcus furiosus
J.Mol.Biol., 293, 1999
|
|
4IJC
 
 | | Crystal structure of arabinose dehydrogenase Ara1 from Saccharomyces cerevisiae | | Descriptor: | D-arabinose dehydrogenase [NAD(P)+] heavy chain, GLYCEROL, SULFATE ION | | Authors: | Hu, X.Q, Guo, P.C, Li, W.F, Zhou, C.Z. | | Deposit date: | 2012-12-21 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Saccharomyces cerevisiaeD-arabinose dehydrogenase Ara1 and its complex with NADPH: implications for cofactor-assisted substrate recognition
Acta Crystallogr.,Sect.F, 69, 2013
|
|
2WQ3
 
 | |
3NAU
 
 | | Crystal structure of ZHX2 HD2 (zinc-fingers and homeoboxes protein 2, homeodomain 2) | | Descriptor: | SULFATE ION, Zinc fingers and homeoboxes protein 2 | | Authors: | Ren, J, Bird, L.E, Owens, R.J, Stammers, D.K, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2010-06-02 | | Release date: | 2010-07-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Novel structural features in two ZHX homeodomains derived from a systematic study of single and multiple domains
Bmc Struct.Biol., 10, 2010
|
|
1BXS
 
 | | SHEEP LIVER CLASS 1 ALDEHYDE DEHYDROGENASE WITH NAD BOUND | | Descriptor: | ALDEHYDE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Moore, S.A, Baker, H.M, Blythe, T.J, Kitson, K.E, Kitson, T.M, Baker, E.N. | | Deposit date: | 1998-10-08 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Sheep liver cytosolic aldehyde dehydrogenase: the structure reveals the basis for the retinal specificity of class 1 aldehyde dehydrogenases.
Structure, 6, 1998
|
|
4G4B
 
 | | Room temperature X-ray diffraction study of cisplatin binding to HEWL in DMSO media with NAG after 7 months of crystal storage | | Descriptor: | CHLORIDE ION, Cisplatin, DIMETHYL SULFOXIDE, ... | | Authors: | Tanley, S.W.M, Schreurs, A.M.M, Kroon-Batenburg, L.M.J, Helliwell, J.R. | | Deposit date: | 2012-07-16 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Room-temperature X-ray diffraction studies of cisplatin and carboplatin binding to His15 of HEWL after prolonged chemical exposure.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1R9X
 
 | | Bacterial cytosine deaminase D314G mutant. | | Descriptor: | Cytosine deaminase, FE (III) ION, GLYCEROL, ... | | Authors: | Mahan, S.D, Ireton, G.C, Stoddard, B.L, Black, M.E. | | Deposit date: | 2003-10-31 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy.
Protein Eng.Des.Sel., 17, 2004
|
|
